
Transcriptomic analysis of feeder-free culture system for maintaining naïve-state pluripotency in human pluripotent stem cells
Introduction
A well-designed medium containing a basic fibroblast growth factor (bFGF), maintains flat monolayer colonies of primed human induced pluripotent stem cells (hiPSCs) in feeder-independent culture conditions (1). Various differentiated cells are derived from hiPSCs for practical use. However, the variability in their degree of pluripotency and differentiation propensities, which depends on the inductive methods and culture conditions, hinders the supply of cells (2). The development of various kinds of pluripotent stem cells (PSCs), in addition to these bFGF-dependent hiPSCs, is necessary to ensure supply of various differentiated cells and to minimize variability. Leukaemia inhibitory factor (LIF)-dependent naïve-like stem cells are derived from mouse embryonic stem cells (ESCs). These cells, characterized by dome-shaped colonies, were first developed in 2013 by adding several small molecule inhibitors in addition to growth factors, such as bFGF and LIF (3). Specific culture conditions for converting primed hiPSCs and human ESCs to LIF-dependent naïve-like PSCs are standardized (4,5). Supplementation with small molecule inhibitors for transgene-independent protocols is used to establish culture conditions for clinical applications. A combination of four or five inhibitors, LIF, and activin A (4i or 5i/L/A) (6) is used in premade commercial media. However, these protocols require feeder cells, such as mouse embryonic fibroblasts (MEFs), to maintain pluripotency. A feeder-free culture method and commercial medium is available (7); however, it requires Matrigel matrix-coated dishes. Matrigel is a biological product de-rived from mouse sarcoma; therefore, it is desirable to exclude it for culturing naïve-like PSCs for clinical applications. Both feeder cells and Matrigel contain animal-derived components and are difficult to handle. Therefore, we aimed to culture human naïve-like PSCs using recombinant vitronectin-coated dishes. In addition to avoiding the use of biological materials, this method enables easy handling.
To enhance pluripotency characteristics, we focused on small-molecule inhibitors that were associated with reprogramming, because reprogramming is closely related to the naïve state (8,9). Pharmacological inhibition of the NOTCH signaling pathway [dibenzazepine (DBZ)] and the use of the histone H3 methyltransferase disruptor of telomeric silencing 1-like inhibitor (DOT1L; iDOT1L) enhanced the reset process of hiPSCs (10). We obtained naïve-like PSCs without hypoxic conditions. The combination of DBZ, a representative γ-secretase inhibitor, and iDOT1L promoted the conversion from the primed to naïve state and the maintenance of the naïve state. Therefore, a combination of growth factors, small-molecule inhibitors, and culture conditions were assessed.
Methods
Ethics statement
Human endometrial tissues from menstrual blood and placental tissues were collected after obtaining signed informed consent. The experiments were approved by the Institutional Review Board of the National Center for Child Health and Development (NCCHD) of Japan (Permit Nos. #146 and #518). All experiments using human cells and tissues were performed in accordance with the tenets of the Declaration of Helsinki (as revised in 2013).
The animal use protocol was approved by the Institutional Animal Care and Use Committee of the NCCHD (Permit No. A2003-002). All animal experiments were based on the three Rs (refine, reduce, and replace), and all efforts were made to minimize animal discomfort and to reduce the number of animals used.
Conventional primed hiPSC culture
All cell lines were maintained in a humidified atmosphere at 37 °C with 5% CO2 in air (21% O2, 5% CO2). We used three types of previously established hiPSCs: cells from menstrual blood (EDOM), cells from the human skin fibroblast (HSF) derived cells, and human adipose tissue-derived stem cells (ADSCs; female, PT5006; Lonza Bioscience, Walkersville, MD, USA). EDOM-iPS cells (EDOM#6-iPSCs) were generated using a Cyto-Tune™-iPS Sendai Reprogramming Kit (Thermo Fisher Scientific, Waltham, MA, USA). The other lines (ADSC-A5 and HSF02) were established through direct delivery of mRNAs encoding reprogramming factors (Stemgent StemRNA 3rd Gen Reprogramming Kit; Reprocell, Yokohama, Japan) and cultured in StemFlex™ Medium (Thermo Fisher Scientific) on vitronectin (Thermo Fisher Scientific, for EDOM#6 and ADSC-A5) (10) or iMatrix-511 (Nippi, Tokyo, Japan, for HSF02). Cells were passaged every 5–7 days by dissociation with Accutase (Thermo Fisher Scientific), and replated onto a matrix coated plate with a ROCK inhibitor (Y-27632; FUJIFILM Wako Pure Chemical, Tokyo, Japan) at a final concentration of 10 µM. These cells were replated at a density of 2×103 cells/cm2. The medium was replaced completely every day, except on the day after passage.
Feeder-free dome-shaped induced pluripotent stem cells (FFDS-iPSCs) culture
FFDS-iPSCs were converted from primed-state hiPSCs and cultured on vitronectin (4×104 cells per cm2). We used a combination of two commercial products, StemFlex™ Medium (Thermo Fisher Scientific) as the basal medium and the supplemental component RSeT™ Medium (RSeT™ 5X Supplement, STEMCELL Technologies, Vancouver, BC, Canada) to supply a combination of inhibitors, LIF, and activin A (4). We combined StemFlex™ Medium (Thermo Fisher Scientific) and RSeT™ 5X Supplement (STEMCELL Technologies) at a ratio of 4:1; 2 µM DBZ (R&D Systems, Minneapolis, MN, USA) and 3 µM iDOT1L (Abcam, Cambridge, UK) were added to this mixed medium for maintaining naïve-state cells under feeder-free conditions. Dimethyl sulfoxide (1:1,000 dilution; Sigma-Aldrich, Missouri, USA) was included as the solvent for the two compounds. At the initiation of FFDS-iPSCs culture, single cells derived from primed hiPSCs were passaged on vitronectin-coated culture plates at a density of 4×104 cells/cm2. Every 2–5 days, all differentiating FFDS-iPSCs were passaged through dissociation with Accutase and replating on vitronectin-coated plates (2×104 cells per cm2) with 10 µM ROCK inhibitor for 48 h. The medium was replaced completely on alternate days. The cell growth rate and doubling time of FFDS-iPSCs (ADSCs) were calculated based on the number of replated cells, the total increase in the number of cells, and the duration of culture. Cell proliferation curves were obtained, and population doubling (PD) at the nth passage (Pn) was determined as follows: PD at Pn = PD at Pn 1 + log2 (counted cells/seeded cells).
Embryoid body (EB) formation for in vitro differentiation assay
Single cells were dissociated and seeded at a density of 1×104 cells in 100 µL medium per well, in uncoated 96-well ultra-low attachment plates (Corning Ultra-Low Attachment Multiple Well Plate; Merck Millipore, Burlington, MA, USA) to form EBs. After 9 days, the EBs were transferred to gelatine-coated tissue culture dishes at a density of approximately three EBs per cm2 and allowed to differentiate further for 3 weeks. The original medium contained 90% DMEM/Nutrient Mixture F-12 (DMEM/F-12), 10% fetal bovine serum (FBS), 1 mM sodium pyruvate, 2 mM L-glutamine, 100 U/mL penicillin, 100 µg/mL streptomycin, and 0.1 mM nonessential amino acids (all reagents from Thermo Fisher Scientific). This medium (30 µL per well) was added at five days after seeding in 96-well plates; the medium was replaced thrice every week.
Teratoma formation for the in vivo differentiation assay
Approximately 1–5×106 cells were injected into the subcutaneous tissue and kidney capsule of nude mice (BALB/cAJcl-nu/nu; CLEA Japan, Tokyo, Japan); tumor masses were collected after 2 to 3 months (11). The harvested tumours were fixed with 4% paraformaldehyde, embedded in paraffin, cut into 5 µm sections, and stained with haematoxylin and eosin. Various parts of the tumours were subjected to histological analysis; the three germ layers were identified based on representative histological features (11).
Immunofluorescence staining
Cells were cultured in a glass-bottom dish (AGC Techno Glass, Shizuoka, Japan) and fixed with 4% paraformaldehyde for 10 min at 4 °C before being permeabilized with 0.1% Triton X-100 (Sigma-Aldrich) for 10 min at room temperature (RT). The samples were blocked with 5% normal goat serum in Dulbecco’s phosphate-buffered saline (DPBS; Thermo Fisher Scientific) for 30 min at RT. They were incubated with primary antibodies at 4 °C overnight. After washing with DPBS, the samples were incubated with secondary antibodies conjugated to Alexa 488 or 546 (Thermo Fisher Scientific) for 30 min at RT. After washing with DPBS, the samples were placed in mounting medium with 4',6-diamidino-2-phenylindole (DAPI). The following primary antibodies were used: OCT4 (C-10; Santa Cruz Biotechnology, Dallas, TX, USA), NANOG (ReproCell), Tra1-60 (MAB4360; Sigma-Aldrich), SSEA4 (MAB1435; R&D Systems), SSEA1 (MAB2155; R&D Systems), KLF4 (ab216875; Abcam), PRDM14 (ab187881; Abcam), anti-βIII Tubulin (TUJ1, Promega, Madison, Wisconsin, USA), α-smooth muscle actin (α-SMA; A2547; Sigma-Aldrich), and SOX17 (MAB1924; R&D Systems). Images were acquired using an LSM510 laser scanning confocal microscope (Carl Zeiss, Oberkochen, Germany). All antibodies except for the anti-TUJ1 antibody (1:300) were used at a 1:150 dilution in 5% normal goat serum.
Quantitative reverse transcription polymerase chain reaction (PCR)
Total RNA from the cell pellet was extracted using ISOGEN II (Nippon Gene, Tokyo, Japan). cDNA was prepared using Superscript® IV Reverse Transcriptase (Thermo Fisher Scientific), according to the manufacturer’s instructions. Gene expression was analysed using Qiagen RT2 Profiler PCR Arrays (Qiagen, Hilden, Germany), which were commercially produced for simultaneously and reliably assessing the expression of genes in various pathways. Total RNA (500 ng) was used with a PCR array kit; PCR was performed using the SYBR Green method (RT2 SYBR® Green qPCR Mastermixes; Qiagen) in a 7300 Real-Time PCR System (Thermo Fisher Scientific), following the manufacturer’s instructions. Threshold cycle values were converted to relative values. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the internal control; the expression level in each sample was normalized to that in a primed ADSC iPS cell line. Representative mRNA levels of pluripotency-related genes in 12 cell lines, including six FFDS-iPSCs lines and six primed hiPSC lines, were assessed with RT2 Profiler™ PCR Array Human Induced Pluripotent Stem Cells (#PAHS-092Z; Qiagen).
Karyotypic analysis
Chromosomal G-band analyses of primed hiPSCs and FFDS-iPSCs were performed at Chromosome Science Labo. Ltd. (Sapporo, Hokkaido, Japan). At least 20 metaphase spreads were examined for each cell line.
Statistical analysis
Quantitative data are reported as the mean ± standard error of the mean (SEM) from at least three independent experiments. ExAtlas (https://lgsun.irp.nia.nih.gov/exatlas/) software for gene expression meta-analysis was used to construct heatmaps and calculate P values. A P value <0.05 was considered statistically significant by Student’s t-test.
Results
Stable cultivation of FFDS-iPSCs
Three independent primed hiPSC lines (ADSC-iPSCs, EDOM-iPSCs and HSF-iPSCs) were used (10). We developed a culture condition for the maintenance of pluripotency of naïve-like hiPSCs under feeder-free conditions. The hiPSCs culture condition contains a NOTCH inhibitor, DBZ, and a histone methyltransferase DOT1L inhibitor (iDOT1L), which we have previously reported in maintaining the naïve state using feeder cells (10). We applied these conditions to culture feeder-free naïve human pluripotent stem cells (hPSCs). However, we were unable to maintain the conversion of naïve hiPSCs from primed hPSCs using the RSeT™ medium supplemented with DBZ and iDOT1L in initial experiments (data not shown). Furthermore, applying another culture condition using the StemFlex™ medium as the basal medium supplemented with LIF and 6 inhibitors, MEK inhibitor, GSK-3b inhibitor, BRAF inhibitor, SRC inhibitor, DBZ, and iDOT1L was unstable in the naïve state (data not shown). We eventually optimized a specific combination of the human PSC culture products supplemented with DBZ and iDOT1L. All investigated PSC lines were maintained as dome-shaped colonies characteristic of naïve-like stem cells over ten passages. Therefore, the culture of the ADSC-iPS cell line was further evaluated (Figure 1A,1B). A cellular proliferation assay showed that FFDS ADSC-iPSCs (FFDS-ADSCs) had a PD time similar to that of primed ADSC-hiPSCs (primed-ADSCs) (FFDS-ADSCs: 1.05±0.27 vs. primed-ADSCs: 0.91±0.22 days, P=0.16; Figure 1C).
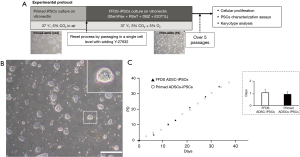
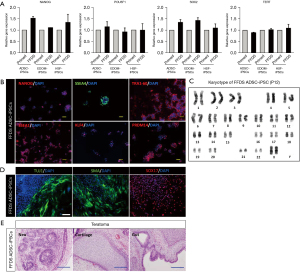
Pluripotency markers and karyotype evaluation of feeder-free iPSCs
To confirm the pluripotency of FFDS-iPSCs, qRT-PCR analysis and immunofluorescence staining for representative pluripotency markers were performed. The transcript levels of the pluripotency markers (NANOG, POU5F1, SOX2 and TERT) in the FFDS-iPSCs were equal to that in primed-hiPSCs (Figure 2A). In addition, immunofluorescence staining indicated NANOG expression in the nuclei of FFDS-ADSCs (Figure 2B); SSEA4 and Tra1-60 were detected in the cytoplasm. In addition, we detected representative naïve pluripotency-related proteins, SSEA1, in the cytoplasm and KLF4 and PRDM14, in the nuclei (Figure 2B). Culture systems used for producing naïve-like stem cells indicated a tendency towards inducing aneuploidy (6,12,13). Therefore, metaphase chromosome analyses with G-banding were performed on the FFDS-iPSCs at passage 12 and displayed normal karyotypes in FFDS-iPSCs (Figure 2C). The pluripotency of the established FFDS-iPSCs was evaluated based on EB formation during in vitro differentiation and teratoma formation in vivo. EBs differentiated from FFDS-iPSCs expressed markers associated with the three germ layers: TUJ1 (ectoderm), alpha-smooth muscle cell (αSMA) (mesoderm), and SOX17 (endoderm; Figure 2D). In the in vivo analysis, representative ectoderm, mesoderm, and endoderm derivatives were detected in all transplant experiments with FFDS-iPSCs (Figure 2E).
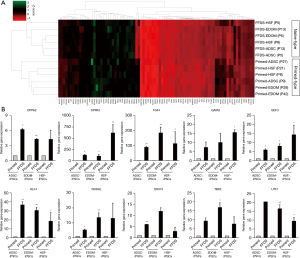
Naïve characterization of FFDS-iPSCs at transcription level
To examine the differences in gene expression between FFDS-iPSCs and primed-iPSCs, we used RT-PCR array kits in which the major transcription factors associated with pluripotency and various differentiation-related genes were included. A total of 84 genes were used for cluster analysis, and ExAtlas (https://lgsun.irp.nia.nih.gov/exatlas/) gene expression meta-analysis software was used for constructing heatmaps. In total, 12 samples, six primed-iPSC and six FFDS-iPSC samples, were used. Hierarchical clustering classified the 12 samples into two groups based on culture conditions (Figure 3A). Based on earlier studies that systematically assessed the characteristics of naïve-like PSCs (4,7,14-16), the well-known naïve pluripotency-related markers, such as DPPA2, DPPA3, FGF4, GATA2, GDF3, KLF4, NODAL, SOX15, TBX3, and UTF1, tended to be highly expressed in FFDS-iPSCs (Figure 3B).
Discussion
Since the first report of human naïve PSCs (3-5), the insights into the hierarchy of human PSCs have evolved. Regarding application for differential works, especially early human developmental, culturing PSCs under naive conditions are becoming indispensable models (17). The latest advancements in generating human embryo-like 3D structures including the blastocyst-like structure, known as blastoid, require naïve state PSCs (18-21). However, there is a lack of standardized culture protocols for human naïve PSCs (22). Further studies for improving the culture conditions of human naïve PSCs are necessary, because they could be aided modelling early human development (17).
We aimed to develop a more convenient and stable cultivation system for human naïve PSCs in a feeder-free culture, to further facilitate human stem cell-based embryology. We evaluated representative culture conditions for naïve-like PSCs, when selecting growth factors and inhibitors (4,6). We used a most popular commercial product, containing a combination of four or five inhibitors, LIF, and Activin A for performing the reset process. Several applications require maintaining naïve state PSCs without feeders; therefore, we added inhibitors of the NOTCH signaling pathway (DBZ) and histone H3 methyltransferase (iDOT1L), to promote pluripotency (8,10). However, we could not maintain dome-shaped colonies in this system. Therefore, we evaluated the supplementation of two growth factors, bFGF and LIF. We combined two commercial products, a base medium for primed hiPSCs and a supplement for naïve-like PSCs. We obtained FFDS-iPSCs, which could be maintained for over 10 passages. FFDS-iPSCs could not be maintained without DBZ and iDOT1L; therefore, these two inhibitors were necessary for stably maintaining the naïve state under feeder-free conditions.
The pluripotency of the native-state iPSCs, FFDS-iPSCs, was verified in both in vitro and in vivo differentiation into the three germ layers; this pluripotency was maintained for a certain culture period. A normal karyotype was retained after 12 passages. The expression of the naïve markers was consistently higher in all three cell lines of FFDS-iPSCs, compared to that in the primed iPSCs. Characterization of the FFDS-iPSCs showed that culturing of human naïve PSCs under refined conditions with feeder-free could facilitate applications using naïve hPSCs.
Conclusions
Combining two growth factors, bFGF and LIF, and further supplementing the medium with DBZ and iDOT1L could stably convert and maintain dome-shaped PSCs from primed hiPSCs under feeder-free conditions in vitronectin-coated plates. These FFDS-iPSC had naïve-like characteristics.
Acknowledgments
We would like to thank the members of our laboratory for their helpful discussions. We also thank Minoru Ichinose and Shoko Miyajima for preparing histologic samples.
Funding: This research was supported by the Japan Agency for Medical Research and Development (AMED) (Grant No. 20bk0104089h0002). This work was supported by the Japan Society for the Promotion of Conventions (No. 201902081cc), the Japan Society for the Promotion of Sciences under Grants-in-Aid for Scientific Research (A) (No. 20H00550) and grant from the National Center for Child Health and Development (No. 30-4).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://sci.amegroups.com/article/view/10.21037/sci-2022-043/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. Human endometrial tissues from menstrual blood and placental tissues were collected after obtaining signed informed consent. The experiments were approved by the Institutional Review Board of the National Center for Child Health and Development (NCCHD) of Japan (Permit Nos. #146 and #518). All experiments using human cells and tissues were performed in accordance with the tenets of the Declaration of Helsinki (as revised in 2013). The animal use protocol was approved by the Institutional Animal Care and Use Committee of the NCCHD (Permit No. A2003-002). All animal experiments were based on the three Rs (refine, reduce, and replace), and all efforts were made to minimize animal discomfort and to reduce the number of animals used.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Chen G, Gulbranson DR, Hou Z, et al. Chemically defined conditions for human iPSC derivation and culture. Nat Methods 2011;8:424-9. [Crossref] [PubMed]
- Kim K, Doi A, Wen B, et al. Epigenetic memory in induced pluripotent stem cells. Nature 2010;467:285-90. [Crossref] [PubMed]
- Gafni O, Weinberger L, Mansour AA, et al. Derivation of novel human ground state naive pluripotent stem cells. Nature 2013;504:282-6. [Crossref] [PubMed]
- Theunissen TW, Powell BE, Wang H, et al. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell 2014;15:471-87. [Crossref] [PubMed]
- Takashima Y, Guo G, Loos R, et al. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell 2014;158:1254-69. Erratum in: Cell 2015;162:152-3. [Crossref] [PubMed]
- Theunissen TW, Friedli M, He Y, et al. Molecular Criteria for Defining the Naive Human Pluripotent State. Cell Stem Cell 2016;19:502-15. [Crossref] [PubMed]
- Szczerbinska I, Gonzales KAU, Cukuroglu E, et al. A Chemically Defined Feeder-free System for the Establishment and Maintenance of the Human Naive Pluripotent State. Stem Cell Reports 2019;13:612-26. [Crossref] [PubMed]
- Ichida JK, Tcw J, Williams LA, et al. Notch inhibition allows oncogene-independent generation of iPS cells. Nat Chem Biol 2014;10:632-9. [Crossref] [PubMed]
- Stuart HT, van Oosten AL, Radzisheuskaya A, et al. NANOG amplifies STAT3 activation and they synergistically induce the naive pluripotent program. Curr Biol 2014;24:340-6. [Crossref] [PubMed]
- Isono W, Kawasaki T, Ichida JK, et al. The combination of dibenzazepine and a DOT1L inhibitor enables a stable maintenance of human naïve-state pluripotency in non-hypoxic conditions. Regen Ther 2020;15:161-8. [Crossref] [PubMed]
- Akutsu H, Nasu M, Morinaga S, et al. In vivo maturation of human embryonic stem cell-derived teratoma over time. Regen Ther 2016;5:31-9. [Crossref] [PubMed]
- Pastor WA, Chen D, Liu W, et al. Naive Human Pluripotent Cells Feature a Methylation Landscape Devoid of Blastocyst or Germline Memory. Cell Stem Cell 2016;18:323-9. [Crossref] [PubMed]
- Sahakyan A, Kim R, Chronis C, et al. Human Naive Pluripotent Stem Cells Model X Chromosome Dampening and X Inactivation. Cell Stem Cell 2017;20:87-101. [Crossref] [PubMed]
- Han X, Chen H, Huang D, et al. Mapping human pluripotent stem cell differentiation pathways using high throughput single-cell RNA-sequencing. Genome Biol 2018;19:47. [Crossref] [PubMed]
- Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell 2009;4:487-92. [Crossref] [PubMed]
- Du P, Pirouz M, Choi J, et al. An Intermediate Pluripotent State Controlled by MicroRNAs Is Required for the Naive-to-Primed Stem Cell Transition. Cell Stem Cell 2018;22:851-864.e5. [Crossref] [PubMed]
- Pera MF, Rossant J. The exploration of pluripotency space: Charting cell state transitions in peri-implantation development. Cell Stem Cell 2021;28:1896-906. [Crossref] [PubMed]
- Liu X, Tan JP, Schröder J, et al. Modelling human blastocysts by reprogramming fibroblasts into iBlastoids. Nature 2021;591:627-32. [Crossref] [PubMed]
- Yu L, Wei Y, Duan J, et al. Blastocyst-like structures generated from human pluripotent stem cells. Nature 2021;591:620-6. [Crossref] [PubMed]
- Fan Y, Min Z, Alsolami S, et al. Generation of human blastocyst-like structures from pluripotent stem cells. Cell Discov 2021;7:81. [Crossref] [PubMed]
- Kagawa H, Javali A, Khoei HH, et al. Human blastoids model blastocyst development and implantation. Nature 2022;601:600-5. [Crossref] [PubMed]
- Semi K, Takashima Y. Pluripotent stem cells for the study of early human embryology. Dev Growth Differ 2021;63:104-15. [Crossref] [PubMed]
Cite this article as: Isono W, Kawasaki T, Ichida JK, Nagasaka K, Hiraike O, Umezawa A, Akutsu H. Transcriptomic analysis of feeder-free culture system for maintaining naïve-state pluripotency in human pluripotent stem cells. Stem Cell Investig 2023;10:10.


