Identification of neural stem cell differentiation repressor complex Pnky-PTBP1
Neural differentiation is a complex process involving numerous cell types and transition programs. Both the embryonic cortical ventricular zone (VZ) and the adult ventricular-subventricular zone (V-SVZ) of the brain contain neural stem cells (NSCs). NSCs are multipotent cells of glial origin capable of giving rise to intermediate progenitor cells that divide before producing migratory young neurons (1). The importance of this process is underscored by the fact that targeting developing neurons locally in specialized brain regions would permit treatment of neurodevelopmental or neurodegenerative diseases in which specific mature neurons are lost or absent (2).
Many molecular factors and pathways govern differentiation programs in all human tissues, organs, and systems. Among these, long non-coding RNAs (lncRNAs) are emerging as critical regulatory molecules in differentiation processes that affect broadly human physiology and pathology (3). LncRNAs are RNA molecules longer than 200 nucleotides lacking apparent protein-coding ability. Thousands of lncRNAs are expressed in the mammalian genome, and they are capable of controlling gene expression programs on many different levels: they can modulate chromatin organization and function, influence gene transcription, alter pre-mRNA metabolism, and affect mRNA turnover and translation (4,5). LncRNAs have been implicated specifically in neural development and differentiation (6). Early examples of this influence include lncRNAs Cyrano and Megamind, which were shown to be necessary for neuronal development, as knockdowns of these lncRNAs decreased the numbers of neurons in zebrafish and mouse embryos, respectively (7). However, lncRNAs playing a role in NSCs and their transition to neurogenic progenitors are only now beginning to emerge (6).
Recently, Ramos and colleagues (8) reported Pnky, a predominantly nuclear and evolutionarily conserved lncRNA expressed only in neural tissues. Expression of Pnky decreased during V-SVZ NSC differentiation into neuronal cells, suggesting that Pnky might be the first known neuronal lncRNA that inhibits neuronal development. This finding contrasts with earlier evidence of other neuronal lncRNAs such as Cyrano and Megamind that instead promoted neuronal development, as revealed when their reduced expression prevented neuronal differentiation [reviewed in (6)]. In contrast, Pnky loss-of-function interventions (e.g., using lentiviral particles with shRNA targeting Pnky) were found to trigger the development of higher numbers of neurons from cultured post-natal V-SVZ NSCs (8). Time-lapse microscopy unexpectedly revealed that suppressing Pnky expression did not increase NSC proliferation, but instead produced more transit-amplifying cells, the intermediate cell type that eventually produces neuroblasts (8). Thus, Pnky knockdown promoted neural lineage largely by decreasing cell death. The authors observed similar results in another model of in vivo neurogenesis. Injection of Pnky shRNA in the embryonic brain of mice led to a relative increase in neurons and decrease in NSCs compared to control shRNA injections, supporting the hypothesis that Pnky represses the production of young neurons in the developing brain (8).
In search for a mechanism that might explain how Pnky elicited this effect, the authors performed affinity pull-down assays using tagged Pnky as RNA bait followed by proteomic analysis to isolate bound proteins. This was a likely approach, as lncRNAs often carry out their function by interacting with RNA-binding proteins (RBPs) (4,5,9,10). Through their association with coding and noncoding RNAs, RBPs have been implicated in all aspects of gene regulation, including nuclear functions like splicing and maturation, and cytoplasmic functions like transport, stability, storage and translation (10,11). RBPs are ubiquitous regulators of cell functions such as division, apoptosis, differentiation, and senescence; however, they are also specialized in the functions of certain tissues, like insulin production, myogenesis, immune cell activation, and adipogenesis. A subset of RBPs is specifically implicated in neural development and in particular in the alternative splicing that leads to isoform diversity (9,12). Numerous studies have uncovered major roles of splicing regulators in the brain such as NOVA and heterogeneous ribonuclear proteins (hnRNPs), including the polypyrimidine tract-binding protein 1 (PTBP1, also known as hnRNP I). PTBP1 was shown to play a role in fibroblast reprogramming to neurons and PTBP1 knockdown led to precocious cortical development (13,14).
Ramos and coworkers identified PTBP1 as a specific binding partner of Pnky in nuclear extracts from V-SVZ NSCs (8). Since nuclear PTBP1 plays a role in controlling splicing, the authors proposed that the complex Pnky-PTBP1 may affect alternative splicing in NSCs leading to neuronal differentiation (Figure 1). In support of this idea were several of the authors’ findings: (I) PTBP1 knockdown in NSCs increased the number of neurons, just as Pnky knockdown increased the number of neurons; (II) many of the same mRNAs were differentially expressed and differentially spliced following PTBP1 knockdown as were following Pnky knockdown; (III) gene ontology analysis revealed that the shared regulated mRNAs encoded proteins with roles in biological processes relevant to neuronal differentiation, such as cell-cell adhesion, synaptogenesis, and neurogenesis.
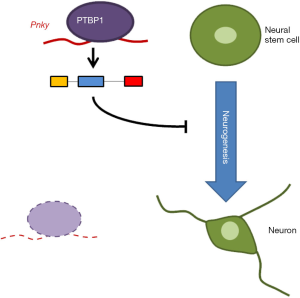
The finding that the Pnky-PTBP1 lncRNP (long noncoding ribonucleoprotein) complex controls splicing during differentiation of NSCs to neurons raises several questions for immediate consideration. It will be important to disrupt PTBP1 or Pnky abundance by alternative methods (e.g., by genomic editing using CRISPR/Cas9 or by alternative silencing RNAs) in order to solidify their observations. It will also be interesting to understand the mechanism whereby this lncRNA-RBP complex modulates splicing, as it could potentially shed light broadly on the field of splicing. For example, does the lncRNA Pnky guide the complex to specific sites of the pre-mRNA, possibly via complementarity with the pre-mRNA, in order for PTBP1 to initiate splicing? Does binding of Pnky to PTBP1 change the affinity of PTBP1 for pre-mRNA sequences?
Among the list of NSC pre-mRNAs selectively spliced by the Pnky-PTBP1 complex, it will be important to identify the actual effectors of Pnky-PTBP1-regulated neurogenesis. In this regard, splicing events that modulate the production of apoptosis regulators might be particularly relevant, given the authors’ finding that silencing Pnky enhanced lineage commitment of NSCs, elevated the number of neurogenic progenitors, and suppressed cell death. Validation that the protein (PTBP1) and the lncRNA (Pnky) components of the complex function jointly to splice specific targets using splicing assays of endogenous and ectopic pre-mRNAs will further strengthen the authors’ model.
The full significance of the interaction of Pnky and PTBP1 also deserves further scrutiny. For example, the Pnky-PTBP1 complex may affect other nuclear functions, such as target mRNA mobilization, maturation, or degradation. Additionally, Pnky likely interacts with other RBPs besides PTBP1, perhaps to regulate subsets of mRNAs outside of those identified in the Ramos report (8) as being coordinately regulated when either PTBP1 or Pnky were silenced. Similarly, PTBP1 associates with other lncRNAs [(15); http://starbase.sysu.edu.cn/)] and these interactions could further influence splicing or other dimensions of gene regulation in NSCs affecting neuronal development.
The significance of the Ramos study (8) is threefold. First, it identifies Pnky as the first lncRNA implicated in controlling neuronal differentiation specifically by modulating the transition of NSCs to intermediate progenitor cells. These results were confirmed in two models of neurogenesis, post-natal and embryonic NSCs. In this process, a critical role is played by the RBP PTBP1. Pnky may also bind to other proteins in order to inhibit neurogenesis.
Second, Pnky is an example of an lncRNA that interacts with a splicing factor to modulate splicing. This model of action contrasts with the well-known example of the splicing-regulatory lncRNA MALAT1, which was shown to function by ‘capturing’ the serine/arginine (SR) splicing factor. By complexing with SR, MALAT1 rendered SR unavailable for endogenous target pre-mRNAs (16,17). By contrast, the Pnky-PTBP1 appears to actively modulate splicing patterns, since losing the lncRNA or the RBP component had similar consequences on gene expression patterns.
Finally, the Ramos report (8) paves the way for the design of possible pharmacological interventions. When targeting specific RNAs or proteins for therapy, it is far easier to silence or inactivate a given molecule that it is to overexpress it in the appropriate tissue, time, and concentration. Therefore, in neurological disease states in which it is desirable to increase the number of differentiated neurons, it may be advantageous to inhibit the Pnky-PTBP1 complex. Although the delivery methods need to improve greatly, one can envision inhibitory therapy directed at shutting off Pnky-PTBP1 activity in order to trigger the progression of NSCs into neurons. Further, since PTBP1 is expressed ubiquitously and in all developmental stages, targeting PTBP1 selectively in neuronal stem cells may be challenging. A more attractive strategy appears to be to target Pnky instead, as its expression is restricted to a small subset of cell types, primarily neuronal stem cells and cells in the subventricular zone and the developing brain. Indeed, in order to design such interventions effectively, it will be important to understand in detail the spatiotemporal pattern of Pnky expression in neural tissue. Such in-depth knowledge could potentially enable exceptionally precise Pnky-directed therapies.
Acknowledgements
This work was supported in full by the National Institute on Aging Intramural Research Program, National Institutes of Health.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Lui JH, Hansen DV, Kriegstein AR. Development and evolution of the human neocortex. Cell 2011;146:18-36. [Crossref] [PubMed]
- Hansen DV, Rubenstein JL, Kriegstein AR. Deriving excitatory neurons of the neocortex from pluripotent stem cells. Neuron 2011;70:645-60. [Crossref] [PubMed]
- Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem 2012;81:145-66. [Crossref] [PubMed]
- Bergmann JH, Spector DL. Long non-coding RNAs: modulators of nuclear structure and function. Curr Opin Cell Biol 2014;26:10-8. [Crossref] [PubMed]
- Yoon JH, Abdelmohsen K, Gorospe M. Posttranscriptional gene regulation by long noncoding RNA. J Mol Biol 2013;425:3723-30. [Crossref] [PubMed]
- Ramos AD, Attenello FJ, Lim DA. Uncovering the roles of long noncoding RNAs in neural development and glioma progression. Neurosci Lett 2015. [Epub ahead of print]. [Crossref] [PubMed]
- Ulitsky I, Shkumatava A, Jan CH, et al. Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 2011;147:1537-50. [Crossref] [PubMed]
- Ramos AD, Andersen RE, Liu SJ, et al. The long noncoding RNA Pnky regulates neuronal differentiation of embryonic and postnatal neural stem cells. Cell Stem Cell 2015;16:439-47. [Crossref] [PubMed]
- Pilaz LJ, Silver DL. Post-transcriptional regulation in corticogenesis: how RNA-binding proteins help build the brain. Wiley Interdiscip Rev RNA 2015;6:501-15. [Crossref] [PubMed]
- Tuck AC, Tollervey D. A transcriptome-wide atlas of RNP composition reveals diverse classes of mRNAs and lncRNAs. Cell 2013;154:996-1009. [Crossref] [PubMed]
- Glisovic T, Bachorik JL, Yong J, et al. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett 2008;582:1977-86. [Crossref] [PubMed]
- Calarco JA, Zhen M, Blencowe BJ. Networking in a global world: establishing functional connections between neural splicing regulators and their target transcripts. RNA 2011;17:775-91. [Crossref] [PubMed]
- Xue Y, Ouyang K, Huang J, et al. Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell 2013;152:82-96. [Crossref] [PubMed]
- Shibasaki T, Tokunaga A, Sakamoto R, et al. PTB deficiency causes the loss of adherens junctions in the dorsal telencephalon and leads to lethal hydrocephalus. Cereb Cortex 2013;23:1824-35. [Crossref] [PubMed]
- Han A, Stoilov P, Linares AJ, et al. De novo prediction of PTBP1 binding and splicing targets reveals unexpected features of its RNA recognition and function. PLoS Comput Biol 2014;10:e1003442. [Crossref] [PubMed]
- Bernard D, Prasanth KV, Tripathi V, et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J 2010;29:3082-93. [Crossref] [PubMed]
- Tripathi V, Ellis JD, Shen Z, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 2010;39:925-38. [Crossref] [PubMed]
Cite this article as: Grammatikakis I, Gorospe M. Identification of neural stem cell differentiation repressor complex Pnky-PTBP1. Stem Cell Investig 2016;3:10.


