
CRISPR-activation-based screen reveals neuronal fate promotion by polycomb repressive complex 2 during direct reprogramming
Conversion of one cell type to another by reprogramming offers valuable opportunities for disease modeling and regenerative medicine. In a much-anticipated scenario, tissues generated from reprogrammed cells will be used to replace degenerated or lost tissues in patients suffering from injury or diseases such as Alzheimer’s, which causes loss of functional nerve cells (neurons) in the brain. Converting the fate of, for instance, a skin cell to a neuron requires the activity of specific transcription factors (TFs). It has previously been demonstrated by the Wernig lab at Stanford University that forced expression of the TFs Brn2, Ascl1, and Myt1l (abbreviated as BAM) in mouse embryonic fibroblasts (MEFs) induces direct reprogramming of MEFs to neurons in a process termed also as transdifferentiation (1). Subsequent studies revealed other TF combinations able to induce direct reprogramming of different source cells to specific types of neurons (2,3).
Recently, a study by Tsunemoto and colleagues identified pairs of basic helix-loop-helix (bHLH) and Pit-Oct-Unc (POU) TFs that promote reprogramming of MEFs to neurons (4). By viral delivery of cDNAs encoding for the respective TFs, they induced their expression and assessed MEF-to-neuron conversion (4). In contrast, Liu and colleagues of the Qi group at Stanford University performed a different strategy to activate expression of TFs and screen for combinations of TFs and DNA-binding factors, which can induce the neuronal fate during differentiation and direct reprogramming (5). They applied CRISPR-activation (CRISPRa), which is a modified version of the CRISPR/Cas9 gene-editing technique and is based on a cleavage domain-deactivated version of the Cas9-protein fused to a transcription activation domain as described previously (6). Such CRISPRa-based induction of endogenous TF expression has been demonstrated previously to sufficiently induce expression of the BAM-TFs, resulting in direct reprogramming of MEFs to neurons (7).
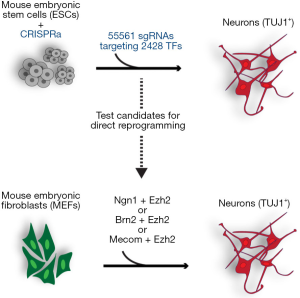
Liu et al. applied the CRISPRa strategy in mouse embryonic stem cells (ESCs) and MEFs to systematically screen for TFs and other DNA-binding proteins that are able to induce differentiation of ESCs to neurons and to directly reprogram MEFs to neuronal cells. The authors first generated a stable mouse ESC line to express CRISPRa components, including a labelled version of the neuron-specific tubulin gene Tubb3 (encoded TUJ1) to generate a visible marker, thereby facilitating the isolation of successfully induced neurons. Next, they created a library containing approximately 55K sgRNAs targeting CRISPRa to 2,428 genes encoding for in silico predicted TFs and DNA binding proteins. Successfully generated neurons via ESC differentiation were isolated by cell sorting, and next generation sequencing was performed to examine which sgRNAs were most efficient in generating neurons. Electrophysiology and transcriptome analysis assessed to which degree the newly generated neurons resemble functional in vivo counterparts. Among the identified 74 targets, several TFs were already known to drive differentiation and reprogramming to neurons. Yet, a number of identified factors, such as Ezh2 (Enhancer of zeste 2), were till then not described to promote neuronal differentiation. Furthermore, the CRISPRa system allowed the assessment of genetic interactions by using two sgRNAs for each target gene, which revealed that the factors Ngn1, Zeb1, Tcf15, Foxo1, Ezh2, Brn2 have mutually positive effects on neuronal differentiation.
Following the CRISPRa-based identification of TFs and DNA-binding proteins that promote neuronal differentiation, the authors tested whether these factors can also induce direct reprogramming to neuronal cells (Figure 1). By viral delivery of DNA constructs to over-express candidate factors in MEFs they assessed MEF-to-neuron direct reprogramming efficiencies. The discovery that over-expression of the Polycomb-Group (PcG) protein-encoding gene Ezh2, in combination with different factors, increased direct reprogramming efficiency, is probably the most interesting finding in this published study. Also, Ezh2 enabled MEF-to-neuron conversion by factors, that alone are not sufficient to induce reprogramming. For instance, Brn2, also known as Pou3f2 (POU domain class 3 transcription factor 2) and Mecom (MDS1 and EVI1 complex locus), cannot induce neuronal direct reprogramming when over-expressed. But in combination with Ezh2, fibroblasts could directly be reprogrammed to neurons. Furthermore, Ezh2 co-expression with the bHLH factor Ngn1 (Neurogenin 1), resulted in direct reprogramming with increased efficiency, boosting the generation of neurons from around 5% (Ngn1 alone) to approximately 50% (Ngn1 + Ezh2). Unlike most other factors identified in this study, Ezh2 is a repressive factor as it is the core subunit of the polycomb repressive complex 2 (PRC2). PRC2 establishes repressive chromatin by mediating methylation of the lysine (K) residue 27 of Histone H3 (H3K27) through the methyltransferase activity of Ezh2 (8). Notably, during ESC to neuron differentiation, Suz12 (suppressor of zeste 12) was also identified—another PRC2 subunit, indicating that PRC2 can have neuron fate-promoting activity. Based on their transcriptome analysis, the authors suggest that Ezh2 might promote the generation of neurons by repressing expression of genes that would otherwise guide cells towards alternative endodermal or mesodermal lineages. Interestingly, the requirement of PRC2 during reprogramming had been made previously by Onder and colleagues during the reprogramming of human fibroblasts to induced pluripotent stem cells (iPSCs) (9). Depletion of Ezh2 or Suz12 by RNA interference significantly reduced reprogramming of fibroblasts to iPSCs, suggesting that PRC2 is required for cell fate switches. Nevertheless, it remains to be determined whether such ex vivo findings prove true in living organisms, as findings in the nematode Caenorhabditis elegans (C. elegans) revealed that PRC2 inhibits reprogramming of germ cells into neurons in vivo (10,11) (Figure 1).
It will be important in the future to reveal the exact mechanism of how PRC2 is selecting genes to repress during neuronal reprogramming. PRC2 lacks DNA-binding domains such as bHLH, POU or Hox domains, which recognize consensus DNA-binding sites in the genome. In Drosophila, PRC2 is recruited to genomic sites by binding to polycomb response elements (PREs) (12). However, the recruitment of PRC2 to specific genes in vertebrate genomes is still not well understood and non-coding RNAs might play a role (13). Further investigation of PRC2 in various biological differentiation and reprogramming systems, primarily in vivo, could reveal context-dependent modes of PRC2-mediated gene repression for promoting either reprogramming or safeguarding cell fates.
Overall, the authors introduced a powerful and versatile strategy by using CRISPRa to identify factors which facilitate the generation of neurons through differentiation and direct reprogramming. It is conceivable that such unbiased approaches to assess the implication of TFs and other DNA-binding factors in differentiation and reprogramming will provide alternative routes to generate specific cell types for tissue engineering, which may help future regenerative medicine applications.
Acknowledgments
We thank Dr. Anna Reid for comments on the manuscript. BT receives funding from ERC-StG-2014-637530 and is supported by the Max Delbrueck Center for Molecular Medicine in the Helmholtz Association.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Vierbuchen T, Ostermeier A, Pang ZP, et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature 2010;463:1035-41. [Crossref] [PubMed]
- Caiazzo M, Dell'Anno MT, Dvoretskova E, et al. Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature 2011;476:224-7. [Crossref] [PubMed]
- Heinrich C, Blum R, Gascón S, et al. Directing astroglia from the cerebral cortex into subtype specific functional neurons. PLoS Biol 2010;8:e1000373. [Crossref] [PubMed]
- Tsunemoto R, Lee S, Szűcs A, et al. Diverse reprogramming codes for neuronal identity. Nature 2018;557:375-80. [Crossref] [PubMed]
- Liu Y, Yu C, Daley TP, et al. CRISPR Activation Screens Systematically Identify Factors that Drive Neuronal Fate and Reprogramming. Cell Stem Cell 2018;23:758-71.e8. [Crossref] [PubMed]
- Mali P, Aach J, Stranges PB, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol 2013;31:833-8. [Crossref] [PubMed]
- Black JB, Adler AF, Wang HG, et al. Targeted Epigenetic Remodeling of Endogenous Loci by CRISPR/Cas9-Based Transcriptional Activators Directly Converts Fibroblasts to Neuronal Cells. Cell Stem Cell 2016;19:406-14. [Crossref] [PubMed]
- Conway E, Healy E, Bracken AP. PRC2 mediated H3K27 methylations in cellular identity and cancer. Curr Opin Cell Biol 2015;37:42-8. [Crossref] [PubMed]
- Onder TT, Kara N, Cherry A, et al. Chromatin-modifying enzymes as modulators of reprogramming. Nature 2012;483:598-602. [Crossref] [PubMed]
- Patel T, Tursun B, Rahe DP, et al. Removal of Polycomb repressive complex 2 makes C. elegans germ cells susceptible to direct conversion into specific somatic cell types. Cell Rep 2012;2:1178-86. [Crossref] [PubMed]
- Seelk S, Adrian-Kalchhauser I, Hargitai B, et al. Increasing Notch signaling antagonizes PRC2-mediated silencing to promote reprograming of germ cells into neurons. Elife 2016. [Crossref] [PubMed]
- Bauer M, Trupke J, Ringrose L. The quest for mammalian Polycomb response elements: are we there yet? Chromosoma 2016;125:471-96. [Crossref] [PubMed]
- Davidovich C, Wang X, Cifuentes-Rojas C, et al. Toward a consensus on the binding specificity and promiscuity of PRC2 for RNA. Mol Cell 2015;57:552-8. [Crossref] [PubMed]
Cite this article as: Wolfram T, Tursun B. CRISPR-activation-based screen reveals neuronal fate promotion by polycomb repressive complex 2 during direct reprogramming. Stem Cell Investig 2019;6:32.


