Signals and transcription factors for specification of human germ cells
Germ cells are the origin of new organisms, since they provide the source of genetic and epigenetic information that is perpetuated from one generation to the next. Germ cells are formed during early embryogenesis and later initiate meiosis giving rise to spermatocytes or oocytes. In humans, primordial germ cell (PGC) development is initiated two weeks after implantation. However, due to ethical and technical constraints, human PGC development has not extensively been investigated.
Our knowledge on germ cell specification is primarily based on observations in the murine system, in which the germ cell fate is initiated in the proximal epiblast around embryonic day 6.5 (E6.5) (1,2). At E7.75 mouse PGCs start migrating along the developing hindgut to the genital ridges, where they arrive at E10.5 (1). During this migration phase PGCs undergo genome-wide DNA demethylation and imprinting erasure, preparing them for meiosis. In 2002, Saitou et al. have started to investigate germ cell competence in mice in more detail by analyzing founder PGCs on single cell resolution (3). The authors demonstrated that upregulation of Fragilis (Ifitm3) marks the onset of germ cell competence, followed by expression of Stella (Dppa3) allowing for retention of pluripotency and escape from the somatic cell fate (3). It was demonstrated later by other groups that PGC fate in mice is regulated by a transcriptional network of BLIMP1, PRDM14 and TFAP2C, which suppresses somatic programs and supports pluripotency (4). The expression of BLIMP1 and PRDM14 is induced by bone morphogenetic protein 4 (BMP4) emerging from the extra-embryonic ectoderm (ExE) in a dose-dependent manner (1). Additionally, WNT3 from the epiblast and visceral endoderm primes the proximal epiblast cells to express Blimp1 by stabilizing SMAD1, a signal transducer of BMP pathways (4). BLIMP1 orchestrates PGC specification by repression of somatic genes, while PRDM14 at the same time supports the pluripotency program (4). TFAP2C (AP-2γ) is located downstream of BLIMP1 and helps in repression of somatic differentiation (4,5).
In vitro, murine PGC-like cells can be established from embryonic stem cells (ESCs) by application of a two-step protocol: ESCs are first incubated with basic fibroblast growth factor (bFGF) and Activin A (ACTA) initiating the formation of Epiblast stem cell like cells (Epi-like cells) (6). Germ cell competence is induced in Epi-like cells under subsequent exposure to BMP4, BMP8B, stem cell factor (SCF), epidermal growth factor (EGF) and leukemia inhibitory factor (LIF) or by direct induction of BLIMP1, PRDM14 and TFAP2C (6). This technique offers a valuable in vitro model for the study of murine PGC development.
The question was, whether the genetic and epigenetic circuitry described in mice would be conserved in other species. Unlike in mice, where PGCs develop from the proximal epiblast, in primates PGCs arise in the nascent amnion as shown for cynomolgus monkeys at E11 (7). Like murine PGCs, cynomolgus PGCs (cyPGCs) express TFAP2C and BLIMP1 at high levels (7). However, cyPGCs display strong expression of SOX17, an endodermal transcriptional regulator that may be regulated by Wnt/β-catenin signaling (7). With respect to their transcriptomic profile (SOX17+, TFAP2C+, BLIMP1+, OCT4+, NANOG+), cyPGCs highly resemble human PGCs (hPGCs). Interestingly, like the murine epiblast, the primate amnion expresses BMP4 and WNT3A (7). This suggested, that cyPGCs share a high degree of similarity to hPGCs in the nascent amnion (7). Thus, the critical role of these signaling molecules for germ cell development seems to be conserved across species and was also suggested to be essential for hPGCs. However, despite their close resemblance to hPGCs, cyPGCs show some interspecies variation, such as CD38 negativity (7). Therefore, the results using cyPGCs still have to be validated in hPGC.
Over the last years, several laboratories have been intensely working on the establishment of human germ cell specification in vitro. In 2015, Irie et al. have described the derivation of human PGC-like cells (hPGCLCs) from human ESCs by sequential application of BMP2 and/or BMP4 using LIF, SCF and EGF (8). These hPGCLCs were characterized by the expression of early germ cell markers (e.g., BLIMP1, TFAP2C) and pluripotency genes (e.g., NANOG, PRDM14), as well as the two endodermal markers GATA4 and SOX17. With respect to their gene expression profile these hPGCLCs closely resemble hPGCs and cyPGCs (7,8). The consecutive expression of BLIMP1 following SOX17 in hPGCLCs suggested that SOX17 acts upstream of BLIMP1, which is in contrast to the murine system where SOX17 is absent and BLIMP1 is a key specifier of PGC fate (4,8). Surprisingly, Irie et al. further showed that SOX17 overexpression alone (w/o BMP2 and/or BMP4, LIF, SCF and EGF) was sufficient to induce hPGCLC fate from hESCs cultured in 4i medium (8). This demonstrates a unique role of SOX17 as key specifier in human germ cell development.
In the same year the group of Saitou published a similar approach whereby hPGCLCs were derived from human induced pluripotent stem cells (hiPSCs) (9). There, hiPSCs were first differentiated into induced mesoderm-like cells (iMeLCs) by application of ACTA and a WNT signaling agonist (CHIR). Subsequently iMeLCs are induced to become hPGCLCs using BMP4, SCF, LIF, EGF and ROCK inhibitor in KSR medium (9). It was discussed, that the iMeLC strategy allows for a more robust and efficient derivation of hPGCLCs when compared to direct hiPSC-hPGCLC conversion (9).
Based on these results, Kobayashi et al. then investigated the induction potential of hPGCs from pluripotent stem cells over the time course of mesendodermal differentiation (10). Peak of competence for hPGC fate was acquired 12 hours during mesendoderm differentiation, at which cells showed moderate expression of primitive streak markers (e.g., EOMES, T) (10). At later time points competence for hPGC fate declined and mesoderm or endoderm fate was induced (10). More detailed analysis using single factor approaches showed that SOX17 alone was able to induce hPGC fate from ESCs, but SOX17 and BLIMP1 together were able to induce hPGC competence more robustly and rapidly (10). Since SOX17 alone drives both PGC and definite endoderm cell fate, BLIMP1 enhances hPGC induction by suppression endoderm genes like FOXA1, FOXA2, HNF1β (10). These findings highlight the role of SOX17 and BLIMP1 as key specifiers for human germ cell competence. However, the previous studies have evaluated the role of SOX17, BLIMP1 and TFAP2C for hPGC induction mostly under artificial settings, i.e., overexpression using a transgene. Transgenic approaches do result in high levels of expression, which are not comparable to physiological levels. For example, the observation that SOX17 alone is able to drive hPGC fate from hESCs has to be viewed critically, since SOX17 levels obtained via ectopic overexpression by far exceed levels found in the in vivo situation.
In a recent publication by the laboratory of Mitinori Saitou, the group has reported a set of very elaborate and elegant experiments to elucidate the role of individual factors for hPGCLC induction. To monitor PGC-fate induction they used a previously established hiPSC line harboring two knock-in reporter constructs: i) Blimp1-TdTomato and ii) TFAP2C-eGF, termed BTAG (9). They applied the system described by Sasaki et al. and Kobayashi et al. where hiPSC were treated with ACTA and CHIR to become iMeLCs which express T, MIXL, and EOMES (9,10). Further treatment of such iMeLCs with BMP4, SCF, EGF and LIF resulted in hPGCLC induction (and upregulation of BTAG) within two days (9,11). BTAG-marker expression could be maintained for at least 6 days (11). In the first experiments, they applied Cas9 mediated gene editing to generate hiPSC cell lines deficient in either SOX17, TFAP2C, BLIMP1, T, MIXL1 or EOMES and monitored the effect of the respective deletion on the formation of hPGCLCs (11).
First, SOX17-deficent hiPSCs were tested, since Irie et al. showed that this factor appears the most upstream factor in hPGCLC formation (8). Expectedly, BT failed to be induced, and AG was only upregulated at day 1, but its expression could not be maintained until day 2 (11). Further to that, SOX17 deficient cells did not induce the PGC surface markers INTEGRINα6 and EpCAM (11). Interestingly SOX17-deficient iMeLCs failed to activate BLIMP1, but induced TFAP2C, NANOS3 and KLF4 and downregulated SOX2 in a normal fashion (11). However, at day two, expression of TFAP2C and NANOS3 could not be maintained (11). So, while SOX17 is required for induction of BLIMP1, induction of TFAP2C and NANOS3 and repression of SOX2 rely at least in part on alternative routes.
Using TFAP2C-deficient hiPSC, at day 2 post iMeLC induction, the rate and the intensity of BT fluorescence was much weaker compared to wildtype cells and the surface markers INTEGRIN 6 and EpCAM were not detectable (11). Interestingly the BT-positive cells expressed lower levels of SOX17, BLIMP1, T, POU5F1 and KLF4 (11). Of note, in BLIMP1-deficient AG-positive cells expression of SOX17 and TFAP2C is not altered (9). So, in humans BLIMP1 seems downstream of TFAP2C.
When looking for a potential signaling network leading to SOX17 activation in human germ cells, Kojima et al. argued that a mesendodermal precursor (iMeLC) state is required for hPGCLC specification (11). This suggested the involvement of one or more factors known to induce the mesendodermal fate. To this end, T, MIXL and EOMES were examined. Neither deficiency of T nor deficiency of MIXL affected the rate and appearance of PGCLCs, suggesting that neither factor is required for PGCLC formation (11). EOMES-deficient clones formed normal iMeLCs and upon induction of PGCLCs showed only moderate induction of AG but not BT (11). INTEGRINα6 and EpCAM could not be detected (11). Here, TFAP2C was induced to a normal level, while SOX17 and BLIMP1 failed to be upregulated (11). So, EOMES is required to activate SOX17 (and BLIMP1) in the course of PGC specification. For further analysis, Kojima et al. generated a doxycycline inducible EOMES allele in the EOMES-deficient hiPSCs. Using this system, they found that continuous induction of EOMES during iMeLC induction did not result in AG activation leading to endodermal differentiation (11). However, this system produced hPGCLC cells expressing key transcription factors at comparable levels to wildtype cells when the induction of EOMES was applied in the latter half of iMeLC induction (11). So, the proper orchestration and timing of EOMES expression (and the other factors) is relevant for PGCLC induction.
Next, the question was addressed whether ACTA and WNT signaling, which are used in iMeLC induction, relate to EOMES during PGCLC induction. PGCLCs were obtained with ACTA and EOMES induction during the latter half of the experiment, but not when CHIR and EOMES were used in the same fashion (11). If ACTA and CHIR were omitted, PGCLC formation was detected on a lower level (11). So, EOMES is required to mediate the WNT signal.
Using the established cell lines, transcriptome data were collected and the differential expressed gene signatures were analyzed. Overlay of the different principal component analyses reveal a very intriguing pattern during the 4-day PGCLC-induction procedure from iMeLCs. Already at day 1 the lack of EOMES and SOX17 results in abrogation of the PGCLC-induction program (11). Deficiency of TFAP2C becomes apparent shortly thereafter, while BLIMP1 deficient cells initiate somatic differentiation as late as day 2 (11). Further mining of the datasets revealed the genes and gene categories which are affected by deletion of each respective factor. This allows differentiating between common functions exerted by all factors and the assignment of specific roles of each factor in the network governing germ cell specification. SOX17, EOMES and TFAP2C all seem to affect the core regulatory circuitry of hPGCs (11). TFAP2C deficient iMeLC did not show any differences to controls, however, in hPGCLCs upregulation of HOX-genes suggestive for mesodermal and neuroectodermal differentiation was detected (11). This suggests that in the human system TFAP2C (and BLIMP1) represses the somatic differentiation program. Further, and to no surprise, there seemed to be a large overlap in the genes affected by loss of SOX17 and EOMES (11). EOMES deficient cells downregulate genes of the WNT pathway, mesoderm development and retinoic acid receptor pathway. Interestingly, EOMES protein was detected in amnion and hypoblast, but not epiblast, of E11 cynomolgus monkey embryos, which at this stage do not show PGC formation (11). These findings support the idea that EOMES functions as key initiator of PGC fate upstream of SOX17 and TFAP2C, by enforcement of a mesoderm intermediate state not only in vitro, but also in vivo.
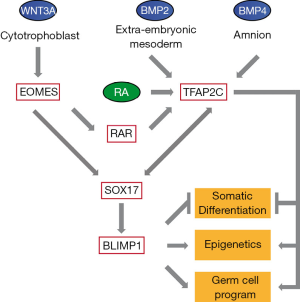
Collectively these data demonstrate that SOX17 and TFAP2C have a pivotal role in specification of human PGCs. WNT signaling emerging from cytotrophoblast leads to expression of the mesendodermal gene EOMES, which transactivates SOX17. SOX17 in turn stimulates expression of BLIMP1. The early amnion and extraembryonic mesoderm are sources of BMP4 and BMP2, respectively, which lead to upregulation of TFAP2C. In addition, TFAP2C was first described as gene upregulated after treatment of P19 embryonal carcinoma cells with retinoic acid (12). Since EOMES leads to upregulation of retinoic acid receptor (RAR) and retinoic acid is present at the time of PGC formation, we hypothesize that TFAP2C expression is induced by BMPs and its expression is maintained by EOMES-mediated induction of the RA-RAR network. Once SOX17, BLIMP1 and TFAP2C are expressed, feedback loops support their own expression. In addition, each factor covers a wide and in part overlapping range of regulatory functions, such as suppression of somatic differentiation and epigenetic machinery and stimulation of the pluripotency network and genes responsible for germ cell development (Figure 1). So, while in vertebrates many of the transcription-factor networks regulating differentiation of somatic cell lineages appear evolutionary conserved, there is considerable variation when looking at the signaling molecules and the transcription factors leading to specification of the germ cell lineage. Applying such in vitro differentiation protocols to further species will shed light on the variation and evolution of the signaling and transcription factor networks in germ cells.
Acknowledgements
The generous funding of the German Science Foundation (DFG) to HS (Scho 503/16-1) is acknowledged.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Saitou M, Yamaji M. Primordial germ cells in mice. Cold Spring Harb Perspect Biol 2012.4. [PubMed]
- Irie N, Tang WWC, Surani MA. Germ cell specification and pluripotency in mammals: a perspective from early embryogenesis. Reprod Med Biol 2014;13:203-15. [Crossref] [PubMed]
- Saitou M, Barton SC, Surani MA. A molecular programme for the specification of germ cell fate in mice. Nature 2002;418:293-300. [Crossref] [PubMed]
- Saitou M, Yamaji M. Germ cell specification in mice: signaling, transcription regulation, and epigenetic consequences. Reproduction 2010;139:931-42. [Crossref] [PubMed]
- Schemmer J, Araúzo-bravo MJ, Haas N, et al. Transcription factor TFAP2C regulates major programs required for murine fetal germ cell maintenance and haploinsufficiency predisposes to teratomas in male mice. PloS One 2013;8:e71113. [Crossref] [PubMed]
- Magnúsdóttir E, Surani MA. How to make a primordial germ cell. Development 2014;141:245-52. [Crossref] [PubMed]
- Sasaki K, Nakamura T, Okamoto I, et al. The germ cell fate of cynomolgus monkeys is specified in the nascent amnion. Dev Cell 2016;39:169-85. [Crossref] [PubMed]
- Irie N, Weinberger L, Tang WWC, et al. SOX17 Is a Critical Specifier of human primordial germ cell fate. Cell 2015;160:253-68. [Crossref] [PubMed]
- Sasaki K, Yokobayashi S, Nakamura T, et al. Robust in vitro induction of human germ cell fate from pluripotent stem cells. Cell Stem Cell 2015;17:178-94. [Crossref] [PubMed]
- Kobayashi T, Zhang H, Tang WWC, et al. Principles of early human development and germ cell program from conserved model systems. Nature 2017;546:416-20. [Crossref] [PubMed]
- Kojima Y, Sasaki K, Yokobayashi S, et al. Evolutionarily Distinctive Transcriptional and Signaling Programs Drive Human Germ Cell Lineage Specification from Pluripotent Stem Cells. Cell Stem Cell 2017;21:517-32.e5. [Crossref] [PubMed]
- Oulad-Abdelghani M, Bouillet P, Chazaud C, et al. AP-2.2: a novel AP-2-related transcription factor induced by retinoic acid during differentiation of P19 embryonal carcinoma cells. Exp Cell Res 1996;225:338-47. [Crossref] [PubMed]
Cite this article as: Jostes S, Schorle H. Signals and transcription factors for specification of human germ cells. Stem Cell Investig 2018;5:13.


