Evolutionarily diverse mechanisms of germline specification among mammals: what about us?
Most of our knowledge on the temporospatial aspects of primordial germ cell (PGC) development in mammalian embryos was obtained through studying laboratory mice (1,2). PGCs are the common precursors of sperm and eggs, and they can be distinguished from their precursors based on their morphological characteristics, migrating activities, and expression of marker molecules. Only recently, information on the cynomolgus monkeys (a.k.a. crab-eating macaque or long-tailed macaque) (3), rhesus monkeys (4), and pigs (5) has been made available. At embryonic day (E) 6.0–6.5 of egg-cylinder-shaped mouse embryos, six PGC precursors are located in the most proximal region of the post-implantation and pre-gastrulation epiblast, close to the extra-embryonic tissues and the region where primitive streak will be formed. A founding population of about 40 PGCs are generated from them at E7.25. The key PGC inducer molecules include bone morphogenetic proteins Bmp2 and Bmp4, which are members of the TGF superfamily growth factors secreted by the visceral endoderm and the extra-embryonic ectoderm, respectively. Extra-embryonic ectoderm also secretes Bmp8b, which restricts development of the anterior visceral endoderm. Because Bmp4 signaling in PGC precursors appears to be antagonized by factors secreted from anterior visceral endoderm, Bmp8b plays a significant role in PGC specification by indirectly enhancing Bmp4 signaling. Wnt3 is required to induce competence for PGC fate in epiblast cells. In contrast to the egg-cylinder shape of mouse embryos around the timing of PGC specification, embryos of pigs, monkeys, and humans show a bilaminar disk morphology. Upon implantation, epiblast of primate embryos segregates amnion to form amniotic cavity and acquire the disk-like shape. Hypoblast gives rise to visceral endoderm and parietal endoderm to form secondary yolk sac. In cynomolgus monkey embryos, PGCs originate from the dorsal amnion at E11 prior to gastrulation, and amnion itself expresses BMP4 and WNT3A. In a large number of phylogenetically diverse placental species including pigs, rabbits, cows, sheep, cats and dogs, epiblast precursor cells polarize to form an epithelial sheet in a manner different from mice and primates (6). Their embryos form inner cell mass and segregate epiblast and hypoblast precursors in a manner similar to mice or primates. However, during the process of epiblast epithelialization, their polar trophoblast degenerates. Consequently, the epiblast disk is directly exposed to the external environment and fuses laterally with mural trophoblast. In porcine embryos, PGC specification occurs within epiblast at posterior, pre-primitive streak component. BMP2, BMP4, and WNT3A are expressed at posterior epiblast around the area of PGC specification. The place where human PGCs were identified in the earliest developmental stage of embryos is on the endoderm wall of yolk sac at the angle with allantois around the end of the third week of gestation (7). This is the same place as migrating mouse PGCs are observed. Not surprisingly, studies tracing back the PGC precursors in pre-gastrulation human embryos have been missing. In particular, little information is available about the inductive process of PGC specification and commitment in vivo.
Then, let us ask this: Are intracellular mechanisms of germline specification largely identical among placental animals? More specifically, can we study mechanisms of germline specification in conventional laboratory animals such as mice and then extrapolate the outcomes to predict human biology? Considering the emerging common scheme that PGC specification in embryos of placental mammals seems to involve signaling of BMP2/4 and WNT3 in epiblast or epiblast-derived cells, this sounds like a reasonable assumption. But recent studies, including one performed by a Kyoto University group led by Mitinori Saitou [Kojima et al. (8)] inform us that things may not be that easy.
Under a circumstance that we even don’t have practical opportunities to access human PGCs and their immediately precursors in embryos for research purposes, how can we study intracellular mechanisms of germline specification? A possible strategy is to study differentiation of human pluripotent stem cells (PSCs) to PGC-like cells (PGCLCs). However, an immediate question to this approach would be this: How precisely does a PGCLC model recapitulate in vivo mechanisms of PGC specification? To address this critique, let us create a grand design. Laboratory mice provide us with convenient opportunities to obtain reasonable amounts of PGCs at various developmental stages, to engineer genes, and to generate functionally certified PSCs—namely, embryonic stem cells (ESCs), epiblast-derived stem cells (EpiSCs), and induced pluripotent stem cells (iPSCs). If we can establish a reasonable level of mechanistic resemblance between specification of mouse embryonic PGCs in vivo and differentiation of mouse PSCs to PGCLCs, then it would be a persuading argument that mechanisms supporting differentiation of human PSCs to PGCLCs may provide useful insight into in vivo mechanisms of human PGC specification.
Three transcription factors Prdm1 (a.k.a. Blimp1), Prdm14, and Tfap2c (a.k.a. Ap2) play critical roles in specification of mouse PGCs (1,9). In the initial engagement phase of mouse PGC specification, both Prdm1 and Prdm14 are induced in PGC precursor epiblast cells by Bmp4. Prdm1 binds to promoters of somatic genes, while Prdm14 binds to distal regulatory elements of somatic genes. Prdm1 and Prdm14 together act as transcriptional suppressors of somatic genes, whereas Prdm14 alone activates germ cell genes. In the next phase, Prdm1 alone induces expression of Tfap2c as a transcriptional activator. In the following, locking-in phase of PGC specification, Prdm14 and Tfap2c together bind to distal regulatory elements of somatic genes, and Prdm1 and Tfap2c also bind together to promoters of somatic genes, overall ensuring transcriptional suppression of somatic differentiation program. On the other hand, Prdm1 and Tfap2c together activate germ cell genes. Thus, a tripartite transcription factor network involving Prdm1, Prdm14, and Tfap2c is critical for mouse PGC specification.
Specification of mouse PGCs occurs within epiblast. All cells in epiblast explants isolated from E5.5-E6.0 mouse embryos are competent to express Prdm1 and Prdm14 in response to Bmp4. When transplanted into seminiferous tubules of neonatal mice, PGCLCs induced from mouse epiblast ex vivo are capable of producing fertilization-competent sperm. Mouse EpiSCs are derived from E5.5–E6.5 post-implantation epiblast and has primed pluripotency, which is in contrast to mouse ESCs derived from inner cell mass of E3.5–E4.5 pre-implantation blastocysts and has naïve pluripotency (10). However, it turned out that neither EpiSCs nor ESCs can directly give rise to PGCLCs by exposure to known PGC-inducing growth factors. In 2011, Saitou’s group cleared this hurdle by showing that male mouse epiblast-like cell culture (EpiLCs—not EpiSCs) robustly generates PGCLCs that are competent to produce fertilization-competent sperm upon transplantation into mouse seminiferous tubules (11). EpiLCs were generated by exposing mouse ESCs or iPSCs to activin A, basic fibroblast growth factor (bFGF), and knockout serum replacement (KSR) in the N2B27 medium. Importantly, EpiLCs gain competence to generate PGCLCs only transiently. Whereas cell aggregates derived from EpiLCs harvested at the second day of EpiLC induction robustly gave rise to PGCLCs upon exposure to BMP4, stem cell factor (SCF), leukemia inhibitory factor (LIF), and epidermal growth factor (EGF) with ~13.5% yield, EpiLCs harvested at the first or third day of induction did not. The same group later showed competence of female mouse PGCLCs thus produced to produce fertilization-competent eggs (12). After this breakthrough, many laboratories have contributed to detailed characterization of mouse PGCLCs, which resemble migrating mouse embryonic PGCs with robustly diminished global genomic DNA methylation involving varying degrees of epigenetic erasure at the imprinting control regions (13-15). A number of studies collectively agreed that mouse PGCLCs faithfully recapitulate the roles of the tripartite transcription factor network in PGC specification.
Robust production of human PGCLCs from PSCs had to wait for 4 years until a report was published in 2015 by a research group lead by Jacob Hanna and Azim Surani (16). Whereas pluripotency of mouse ESCs and iPSCs is in a naïve state, human ESCs and iPSCs in the conventional cell culture conditions are in a primed state, similar to mouse EpiSCs (10). In 2013, Hanna’s group published the first report of derivating naïve pluripotency human ESCs and iPSCs using the naïve human stem cell medium (NHSM) containing chemical inhibitors to protein kinases ERK1/2, GSK3, JNK, ROCK, PKC, and p38 MAPK as well as growth factors LIF, TGF, bFGF (17). Taking advantage of this naïve pluripotency human PSCs, Hanna and Surani developed a protocol that robustly produces human PGCLCs (16). Following this second major breakthrough, several other laboratories (including that of Wolf Reik, Hans Schöler, and ours) reported generations of human PGCLCs from PSCs using slightly different protocols (14,18-20), and a study reported by Mitsunaga et al. provided evidence that human PGCLCs generated using different protocols are transcriptionally similar to each other (20). Available evidence supports the resemblance of human PGCLCs to early-stage human embryonic PGCs prior to the robust epigenetic erasure (14) and/or chemotactic migration (20).
The significant differences in mechanisms of PGC specification between mice and humans soon became evident. The report of Hanna and Surani already recognized SOX17 as a critical upstream inducer of PRDM1 during specification of human PGCLCs (16). This was a surprising finding because in mouse embryos Sox17 is involved exclusively in endodermal differentiation with no known roles in germline specification. During human PGCLC specification, PRDM1 represses a default program of neuronal differentiation while stabilizing transcription of germline genes (18). In mouse embryos, a T-box transcription factor Brachyury (a.k.a. “T”) is induced at primitive streak by Wnt signaling and play critical roles in PGC specification by directly activating Prdm1 and Prdm14 (21). A second T-box transcription factor Eomes (eomesodermin) is also expressed in proximal epiblast prior to primitive streak formation. During gastrulation, expression of Eomes in mouse embryos is restricted to primitive streak. No evidence supports significant roles of Eomes in mouse PGC specification. Roles of BRACHYURY and EOMES in human PGCLC specification were unknown until the report by Kojima et al. (8).
Now that a convenient reliable cell culture model of human PGC specification is available and detailed gene expression schedules during gametogenesis in human and mouse embryos are established (22), Kojima et al. performed systematic CRISPR/Cas9 gene knockout study to determine functional importance of six transcription factors—namely, SOX17, TFAP2C, PRDM1, BRACHYURY, MIXL, and EOMES. The authors used a mutated Cas9 nuclease that cuts only one of the two DNA strands (which is commonly referred to as a “nickase”) with a pair of guide RNAs to reduce the risk of off-target effects. They examined two independent homozygous null clones that exhibited frameshift nonsense mutations in the two target alleles, and the absence of each target transcription factor was confirmed by immunoblotting. Normal diploid karyotype was confirmed for each clone. With this meticulous and setting, they converted human iPSCs to iMeLCs (incipient mesoderm-like cells) and then attempted to generate PGCLCs from iMeLCs following their previously established protocol (18). Whereas SOX1−/− cells were converted to iMeLCs normally, they failed to produce PGCLCs, lacking induction of PRDM1 as well as losing prolonged expression of NANOG, TFAP2C, and NANOS3. Similarly, conversion of TFAP2C-/- cells to iMeLCs was not affected, but their competence to produce PGCLCs was severely impaired, if not totally lost. Expression of NANOG, BRACHYURY, SOX17, PRDM1, KLF4, and NANOS3 was significantly diminished in the PGCLC-inducing cell culture condition. These observations were consistent with the transcription factor binding motif analysis reported by Mitsunaga et al., which identified the canonical TFAP2C binding motifs within 250 bps upstream of transcription start sites of human BRACHYURY, SOX17, and NANOS3 genes (20). Because PRDM1−/− cells express SOX17 and TFAP2C normally in the same condition (18), the authors concluded that TFAP2C acts upstream of PRDM1, which is in contrast to the case of mice where Tfap2c is induced by Prdm1 (1). Striking difference between mice and human was also evident with BRACHYURY, whose knockout in human iPSCs did not affect production of iMeLCs or PGCLCs at all whereas Brachyury is essential for mouse PGC specification through directly activating Prdm1 and Prdm14 (21). Knockout of MIXL1, which drives expression of Sox17 for endoderm specification in zebrafish, did not affect iMeLC or PGCLC production, either. Instead, knockout of EOMES failed to generate PGCLCs whereas induction of iMeLCs occurred normally. The lack of EOMES caused almost complete absence of SOX17 induction in the PGCLC producing condition, and expression of other genes followed the same pattern of the SOX17−/− cells. Importantly, TFAP2C was again induced normally. Overexpression of EOMES from an inducible vector in the EOMES−/− cells during the PGCLC induction culture efficiently rescued the robust PGCLC production as well as induction of germline genes, including SOX17. In contrast, induced overexpression SOX17 also rescued the robust PGCLC production but without inducing EOMES. Thus, EOMES is a critical upstream inducer of SOX17. By mixing the EOMES−/− cells with their parental, normal human iPSCs, the authors determined that EOMES is required cell autonomously for human PGCLC specification. They further provided immunofluorescence staining data supporting expression of EOMES in E11-E15 cynomolgus monkey embryos at the close proximity to locations of PGC specification. Using 18 human ESC clones, a group led by Amander Clark later confirmed that germline competency of human ESCs is also dependent on expression of EOMES (23). Overall, the study of Kojima et al. provided critical pieces of information to understand similarities and differences in transcriptional mechanisms involved in PGC specification. In mice, WNT signaling induces Brachyury and BMP signaling induces PRDM1. Brachyury induces Prdm1 and Prdm14, and Prdm1 induces Tfap2c. The tripartite transcription factors Prdm1, Prdm14, and Tfap2c together promote the germline specification program. In contrast, in human embryos WNT signaling induces EOMES, which then induces SOX17. SOX17 induces PRDM1 and weakly contributes to TFAP2C induction. BMP signaling is also required to induce SOX17 and TFAP2C. Then, SOX17, PRDM1, and TFAP2C together promote the germline specification program, apparently without strictly requiring PRDM14 (19). An independent study performed by the group of Surani indicates that balanced expression of SOX17 and PRDM1 seems important for specification of human PGCLCs (5).
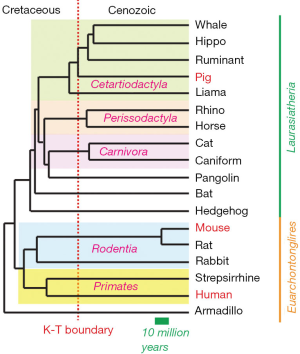
From the viewpoint of phylogenetics, it is interesting that mechanisms of PGC specification in porcine embryos are dependent on SOX17 and, hence appear to be more similar to humans than mice (5), because the phylogenetic distance between pigs and humans seems greater than the distance between mice and humans [see the Figure 1, modified from (24)]. Divergence between primates and rodents occurred about 87–88 million years ago, preceding the abrupt extinction of dinosaurs (the K-T boundary, which was 65 million years ago) by about 22–23 million years. On the other hand, the split between Euarchontoglires (primates, rodents, rabbits, treeshrews and flying lemurs) and Laurasiatheria (most other placental mammals, including pigs) occurred 94 million years ago, significantly preceding the primates-rodents separation. A possibility that mice have been experiencing an exceptionally rapid and reproductively advanced evolution has been recognized, and the laboratory mice rats, and hamsters belong to the most recently evolved rodents—namely, the superfamily Muridae (25). Indeed, embryo of plains vizcacha, a large South American rodent that have longer generation intervals, develops a flat embryonic disc rather than an egg cylinder (https://www.researchgate.net/publication/322851606). It would be interesting to determine whether PGC specification of plains vizcacha follows the mouse pattern (in which Brachyury, Prdm1, Prdm14, and Tfap2c play critical roles) or primate/pig pattern (which is dependent on EOMES, SOX17, PRDM1, TFAP2C).
Acknowledgements
Funding: T Shioda is supported by NIH grants R01ES023316 and R21ES024861.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Magnúsdóttir E, Surani MA. How to make a primordial germ cell. Development 2014;141:245-52. [Crossref] [PubMed]
- Ohinata Y, Ohta H, Shigeta M, et al. A signaling principle for the specification of the germ cell lineage in mice. Cell 2009;137:571-84. [Crossref] [PubMed]
- Sasaki K, Nakamura T, Okamoto I, et al. The Germ Cell Fate of Cynomolgus Monkeys Is Specified in the Nascent Amnion. Dev Cell 2016;39:169-85. [Crossref] [PubMed]
- Clark AT, Gkountela S, Chen D, et al. Primate Primordial Germ Cells Acquire Transplantation Potential by Carnegie Stage 23. Stem Cell Reports 2017;9:329-41. [Crossref] [PubMed]
- Kobayashi T, Zhang H, Tang WWC, et al. Principles of early human development and germ cell program from conserved model systems. Nature 2017;546:416-20. [Crossref] [PubMed]
- Sheng G. Epiblast morphogenesis before gastrulation. Dev Biol 2015;401:17-24. [Crossref] [PubMed]
- De Felici M. Origin, Migration, and Proliferation of Human Primordial Germ Cells. Oogenesis 2012:19-37.
- Kojima Y, Sasaki K, Yokobayashi S, et al. Evolutionarily Distinctive Transcriptional and Signaling Programs Drive Human Germ Cell Lineage Specification from Pluripotent Stem Cells. Cell Stem Cell 2017;21:517-532.e5. [Crossref] [PubMed]
- Magnúsdóttir E, Dietmann S, Murakami K, et al. A tripartite transcription factor network regulates primordial germ cell specification in mice. Nat Cell Biol 2013;15:905-15. [Crossref] [PubMed]
- Weinberger L, Ayyash M, Novershtern N, et al. Dynamic stem cell states: naive to primed pluripotency in rodents and humans. Nat Rev Mol Cell Biol 2016;17:155-69. [Crossref] [PubMed]
- Hayashi K, Ohta H, Kurimoto K, et al. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell 2011;146:519-32. [Crossref] [PubMed]
- Hayashi K, Saitou M. Generation of eggs from mouse embryonic stem cells and induced pluripotent stem cells. Nat Protoc 2013;8:1513-24. [Crossref] [PubMed]
- Shirane K, Kurimoto K, Yabuta Y, et al. Global Landscape and Regulatory Principles of DNA Methylation Reprogramming for Germ Cell Specification by Mouse Pluripotent Stem Cells. Dev Cell 2016;39:87-103. [Crossref] [PubMed]
- von Meyenn F, Berrens RV, Andrews S, et al. Comparative Principles of DNA Methylation Reprogramming during Human and Mouse In Vitro Primordial Germ Cell Specification. Dev Cell 2016;39:104-15. [Crossref] [PubMed]
- Miyoshi N, Stel JM, Shioda K, et al. Erasure of DNA methylation, genomic imprints, and epimutations in a primordial germ-cell model derived from mouse pluripotent stem cells. Proc Natl Acad Sci USA 2016;113:9545-50. [Crossref] [PubMed]
- Irie N, Weinberger L, Tang WW, et al. SOX17 is a critical specifier of human primordial germ cell fate. Cell 2015;160:253-68. [Crossref] [PubMed]
- Gafni O, Weinberger L, Mansour AA, et al. Derivation of novel human ground state naive pluripotent stem cells. Nature 2013;504:282-6. [Crossref] [PubMed]
- Sasaki K, Yokobayashi S, Nakamura T, et al. Robust In Vitro Induction of Human Germ Cell Fate from Pluripotent Stem Cells. Cell Stem Cell 2015;17:178-94. [Crossref] [PubMed]
- Sugawa F, Araúzo-Bravo MJ, Yoon J, et al. Human primordial germ cell commitment in vitro associates with a unique PRDM14 expression profile. EMBO J 2015;34:1009-24. [Crossref] [PubMed]
- Mitsunaga S, Odajima J, Yawata S, et al. Relevance of iPSC-derived human PGC-like cells at the surface of embryoid bodies to prechemotaxis migrating PGCs. Proc Natl Acad Sci USA 2017;114:E9913-E9922. [Crossref] [PubMed]
- Aramaki S, Hayashi K, Kurimoto K, et al. A mesodermal factor, T, specifies mouse germ cell fate by directly activating germline determinants. Dev Cell 2013;27:516-29. [Crossref] [PubMed]
- Saitou M, Miyauchi H. Gametogenesis from Pluripotent Stem Cells. Cell Stem Cell 2016;18:721-35. [Crossref] [PubMed]
- Chen D, Liu W, Lukianchikov A, et al. Germline competency of human embryonic stem cells depends on eomesodermin. Biol Reprod 2017;97:850-61. [Crossref] [PubMed]
- Springer MS, Murphy WJ, Eizirik E, et al. Placental mammal diversification and the Cretaceous-Tertiary boundary. Proc Natl Acad Sci USA 2003;100:1056-61. [Crossref] [PubMed]
- Johnson AD, Alberio R. Primordial germ cells: the first cell lineage or the last cells standing? Development 2015;142:2730-9. [Crossref] [PubMed]
Cite this article as: Mitsunaga S, Shioda T. Evolutionarily diverse mechanisms of germline specification among mammals: what about us? Stem Cell Investig 2018;5:12.


