Targeting chronic myeloid leukemia stem cells: can transcriptional program be a druggable target for cancers?
Introduction
Chronic myeloid leukemia (CML) is a myeloproliferative neoplasm, characterized by clonal expansion and accumulation of differentiated myeloid cells in the blood and the bone marrow (1). CML usually begins with chronic phase, where expanded myeloid cells are almost normal in terms of function. Without effective therapy, most patients progress to accelerated phase and blast phase, as CML progenitors lose terminal differentiation capacity. CML stems from reciprocal translocation t(9;22)(q34;q11), called Philadelphia chromosome, generating constitutively active fusion kinase BCR-ABL. BCR-ABL is a sole oncogenic driver of CML, essentially believed to be sufficient to establish and sustain chronic phase CML (2), while a recent evidence suggests some additional changes may be required for development of CML (3). BCR-ABL confers proliferative and survival advantage to hematopoietic cells by activating several downstream signaling, including rapidly accelerated fibrosarcoma (RAF)/mitogen-activated activated protein kinase (MEK)/extracellular signal-regulated kinase (ERK), Janus kinase (JAK)/signal transducer and activator of transcription (STAT) and phosphatidylinositol 3-kinase (PI3K)/AKT pathways. Cumulative accumulation of chromosomal and molecular changes in addition to BCR-ABL is associated with disease progression to advanced phase diseases, often fetal conditions.
The advent of an orally active small-molecule tyrosine kinase inhibitor (TKI) in 2001 has revolutionized the treatment of CML, marking the start of the era of targeted cancer therapy (4). TKIs can block downstream signals of BCR-ABL effectively and eliminate most CML cells both in vitro and in vivo. In fact, TKIs have changed a fatal disease into controllable disease, with 10-year survival over 80% (5). Most CML patients, however, cannot be cured solely by TKIs and committed to life-long TKI dependence. In fact, around half of the patients who achieved even complete molecular response upon TKI therapy experience CML relapse once they discontinue TKIs (6). Long-term TKI treatment is often associated with adverse effects, acquired resistance and high monetary costs, imposing a substantial burden on healthcare resources as well as individual patients. Furthermore, even under TKI therapy, there remains unignorable risk of progression to advanced stages (7), which cannot be cured without intensive therapy such as allogenic hematopoietic stem cell (HSC) transplantation.
Leukemia stem cells (LSCs)
As many studies using murine retroviral bone marrow transplantation models or xenograft transplantation models have consistently shown, CML is maintained by a very small number of CML LSCs, as is the case of hierarchically-structured normal hematopoietic system, sustained by self-renewing HSCs at the apex of developmental hierarchy (1,8,9). Molecular bases of persisting disease after TKI therapy have been extensively investigated, and many studies have attributed them to LSC persistence even under prolonged and apparently efficacious TKI treatment (10). CML LSCs, unlike their differentiated progenies, don’t depend on BCR-ABL signaling for their survival, in other words, they are free from oncogene addiction (11). Importantly, CML LSCs are very similar to normal HSCs in many aspects, not only indicating the fact that CML LSCs are derived from HSCs, but also suggesting inherent difficulties in selectively eradicating LSCs without sparing HSCs (1,9).
Wnt/β -catenin signaling and its related factors in HSCs and LSCs
For the establishment of targeted therapy against CML LSCs, much effort has been devoted to comprehensively elucidate differences between LSCs and normal HSCs. These studies have illustrated that most key molecules essential for the maintenance of stemness are shared by LSCs and HSCs, including those implicated in transcription, regulation of cell cycle, metabolism, autophagy, signal transduction, and niche-associated factors, while LSCs are more dependent on some of these factors than HSCs, providing potential therapeutic opportunities (1). Among them, a long list of studies have probed into the role of Wnt/β-catenin signaling in normal and malignant hematopoiesis, including the pathogenesis of CML, as depicted in recent reviews (12,13). Wnt/β-catenin signaling is discerned into canonical and non-canonical pathways (Figure 1). In the canonical pathway, so-called destruction complex, consisted of two negative regulatory kinases, degrade β-catenin by targeting it for ubiquitination in the absence of Wnt ligands. After Wnt protein binds to Frizzled family receptor, β-catenin accumulates in the cytoplasm followed by translocation into the nucleus. In the nucleus, β-catenin, not having an ability to bind to DNA directly, modulates gene expression by interacting with members of TCF/LEF transcription factor family; the most extensively studied and widely acknowledged effectors of β-catenin. TCF/LEF factors, consisting of four members: Tcf1, Tcf3, Tcf4 and Lef1, generally act as transcriptional repressors, while binding to β-catenin turns them into transcriptional activators. β-catenin can exert its effects in some contexts through interactions with other transcription factors, including Prop1, forkhead box O (FOXO) factors and Krueppel-like factor 4. As non-canonical Wnt pathways independent of β-catenin, Planar Cell Polarity and Wnt/Ca2+/ nuclear factor of activated T-cells (NFAT) pathways have been well characterized. Furthermore, Wnt signaling is modified by several classes of secreted factors; some inhibit binding of Wnt to its receptors and others isolate its co-receptors, composing multilayered regulation of Wnt activity.
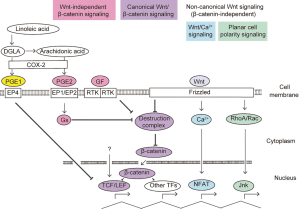
Numerous studies have demonstrated substantial roles of Wnt/β-catenin signaling in the self-renewal of normal HSCs, with some opposing results depending on experimental systems utilized (12). A recent study illustrated that a differential, lineage-specific optimal Wnt dosage within a narrow window is required for robust hematopoietic function, including HSCs, myeloid precursors and T cell precursors (14), suggesting that apparent discrepancies of previous studies can be explained by different levels of Wnt/β-catenin signal activation. In addition, above-mentioned complex and redundant regulation of this pathway at multiple steps might complicate the problem.
In contrast to the conflicting results in the roles of Wnt/β-catenin signaling in normal HSCs, past studies have consistently supported its essential roles in CML, in terms of maintenance of CML LSCs (15,16), TKI resistance (17) and disease progression to blast phase CML (18,19). BCR-ABL directly stabilizes β-catenin through its kinase activity, being an essential process in CML development in mice models (15). Both genetic ablation of β-catenin (catenin beta 1) and pharmacologic inhibition of COX2, which reduces β-catenin level by inhibiting prostaglandin E2 (PGE2) production (20), contribute to decreasing in CML cells resistant to TKIs, indicating β-catenin is also required for the maintenance of TKI-resistant LSC (16). Wnt/Ca2+/NFAT pathway, one of a major component of non-canonical Wnt pathway, is also involved in the survival of CML cells independently of β-catenin, when BCR-ABL is inhibited (21). Constitutive TCF/LEF transcriptional activity in the absence of β-catenin stabilization has been reported in several human hematological tumor cells including K562 cells, a cell line derived from CML, which points to the presence of β-catenin independent activation of TCF1/LEF1 (22). These findings suggest the potential difficulty in targeting Wnt pathways, as well as the complexity of the cellular network regulated by Wnt and related factors in CML.
Tcf1/Lef1 and their associated transcriptional programs as a therapeutic target in LSCs
In 2016, it has been reported that genetic ablation of Tcf1 and Lef1, downstream transcription target of β-catenin, severely compromised CML LSCs, represented by impaired leukemia initiation and maintenance in CML mice models, while normal HSC functions are modestly compromised, which is observed only under the settings of competitive transplantation and regenerative stress (23). In this study, downstream targets of Tcf1 and Lef1 in CML have remained elusive, though functional defects of Tcf1/Lef1-deficient HSCs in hematopoietic reconstitution have been attributed to Egr1 and Tcf3, transcription factors positively regulated by Tcf1 and Lef1. To investigate the molecular background of the differential dependence of LSCs on Tcf1 and Lef1, the same group has performed RNA-sequencing (RNA-seq) analyses comparing between wild-type (WT) and Tcf1/Lef1 double-knockout (DKO) hematopoietic stem and progenitor cells (HSPCs), in addition to corresponding LSCs from CML model mice receiving P210BCR-ABL-transduced bone marrow transplant (24). Cross-comparison of differentially expressed genes between WT and DKO HSPCs and those between WT and DKO LSCs has shown that Tcf1/Lef1 regulates a distinct transcriptional program only in LSCs, with multiple members of activator protein 1 (AP-1) transcription factors enriched in either common or LSC-specific differentially expressed genes. Furthermore, two gene sets, upregulated in human quiescent CML LSCs compared with normal HSPCs, were downregulated in DKO LSCs, suggesting that Tcf1/Lef1 deficiency partly impairs the transcriptional program that maintains LSCs. By using Connectivity Map (CMAP) database, the authors tried to find compounds that can induce gene expression changes recapitulating those caused by Tcf1/Lef1 loss in LSCs (25). Among a couple of candidate compounds, PGE1 significantly impaired colony forming activity of CML LSCs. PGE1 and PGE2 are closely related 20-carbon fatty acid derivatives produced by cyclooxygenase (COX)-dependent oxygenation, of dihomo-γ-linolenic acid (DGLA) for PGE1 and arachidonic acid, a derivative of DGLA, for PGE2. In spite of the structural similarities, their biological functions are strikingly different; PGE1 is characterized by vasodilative effect, usually used to treat conditions such as pulmonary hypertension and peripheral artery occlusive disease, while PGE2 has been involved in more diverse physiological phenomena, including inflammation, bone resorption and parturition, and it can also be utilized for in vitro expansion of human cord-blood HSPCs. In contrast to the fact that pharmacological inhibition of PGE2 signaling is effective against CML through suppressing β-catenin in LSCs (16), the roles of PGE1 in Wnt/β-catenin signaling as well as hematopoietic diseases including CML hadn’t been elucidated. The authors revealed that PGE1, but not PGE2, greatly diminished CML LSCs and prolonged survival of CML mice without affecting the onset of CML. PGE1 abrogated LSCs more effectively than blocking overall PGE production by COX2 inhibitor indomethacin. Furthermore, PGE1 diminished CML LSCs as efficiently as tamoxifen-induced ablation of Tcf1/Lef1, showing a synergistic therapeutic effect upon a TKI, which preferentially affects more differentiated cells.
In an attempt to elucidate mechanistic basis of the anti-CML activity of PGE1, further transcriptomic analyses of DKO and PGE1- and PGE2-stimulated LSCs have been performed, revealing that PGE1 and PGE2 activate distinct pathways and target genes in LSCs; about 26% of downregulated genes in DKO LSCs, including Egr1 and AP-1 factors such as FosB, were repressed by PGE1 but not by PGE2 (24). Ectopic expression of these downregulated AP-1 factors rendered LSCs less sensitive to PGE1, indicating that AP-1 factors are major targets for PGE1-mediated LSC dysfunction. Among four E-prostanoid receptors, designated EP1, EP2, EP3 and EP4, the authors revealed that PGE1 exerts its antileukemic activity through the EP4 receptor independently of β-catenin, by use of Vav1-Cre+/EP4flox/flox and Vav1-Cre+/β-cateninflox/flox LSCs. Misoprostol, a widely-used analog of PGE1, impaired LSC activity in murine primary, secondary and serial transplant CML models, as well as in murine xenograft models using human CML cells. Its activity was again more pronounced in conjugation with TKIs. Importantly, PGE1 was also effective against CML LSCs from patients on advanced stages such as accelerated and blast phase, less sensitive to TKIs, suggesting that PGE1-mediated AP-1 repression is a conserved regulatory pathway in targeting LSCs at different stages of CML.
Future prospects
By targeting a specific transcriptional program that specifically targets CML LSCs, the study by Li et al. (24) provided promising therapeutic approach to LSCs, including those of near intractable advanced disease. Some open questions, however, remains to be solved. Firstly, the whole picture of β-catenin-independent regulation and activity of Tcf1/Lef1 have been still obscure. The difference in the mechanisms and consequences between β-catenin-dependent and -independent activation of Tcf1/Lef1 remain to be clarified, as well as the physiological role of above mentioned PGE1-induced signaling. Li et al. suggested that PGE1-mediated FosB and Fos down-regulation was completely abrogated or even reversed by pharmacologically inhibiting any one of MEK, PI3K and Ca2+, provoking a question how these pathways are interconnected and integrated into β-catenin-independent Tcf1/Lef1 signaling. Secondly, the mechanism how Tcf1/Lef1 play a specific role in CML LSCs and its possible relationship to BCR-ABL and/or other oncogenic signals should be further investigated. Possible functional association of Tcf1/Lef1 with BCR-ABL may lead to a novel therapeutic opportunity for the treatment of Philadelphia chromosome positive acute lymphoblastic leukemia. In addition, because constitutive activation of Wnt/β-catenin signal is a nearly universal features common to many malignancies, clarifying whether Tcf1/Lef1-mediated transcriptional regulation has specific role also in other malignancies, especially in the maintenance of cancer stem cells, might lead to novel therapeutic opportunities.
Acknowledgements
None.
Footnote
Conflicts of Interest: M Kurokawa received the followings: research funding from Pfizer, Novartis and Bristol-Myers Squibb; lecture honoraria from Bristol-Myers Squibb. The other author has no conflicts of interest to declare.
References
- Holyoake TL, Vetrie D. The chronic myeloid leukemia stem cell: stemming the tide of persistence. Blood 2017;129:1595-606. [Crossref] [PubMed]
- Daley GQ, Van Etten RA, Baltimore D. Induction of chronic myelogenous leukemia in mice by the P210bcr/abl gene of the Philadelphia chromosome. Science 1990;247:824-30. [Crossref] [PubMed]
- Foley SB, Hildenbrand ZL, Soyombo AA, et al. Expression of BCR/ABL p210 from a knockin allele enhances bone marrow engraftment without inducing neoplasia. Cell Rep 2013;5:51-60. [Crossref] [PubMed]
- O'Brien SG, Guilhot F, Larson RA, et al. Imatinib compared with interferon and low-dose cytarabine for newly diagnosed chronic-phase chronic myeloid leukemia. N Engl J Med 2003;348:994-1004. [Crossref] [PubMed]
- Hochhaus A, Larson RA, Guilhot F, et al. Long-Term Outcomes of Imatinib Treatment for Chronic Myeloid Leukemia. N Engl J Med 2017;376:917-27. [Crossref] [PubMed]
- Mahon FX, Réa D, Guilhot J, et al. Discontinuation of imatinib in patients with chronic myeloid leukaemia who have maintained complete molecular remission for at least 2 years: the prospective, multicentre Stop Imatinib (STIM) trial. Lancet Oncol 2010;11:1029-35. [Crossref] [PubMed]
- Druker BJ, Guilhot F, O'Brien SG, et al. Five-year follow-up of patients receiving imatinib for chronic myeloid leukemia. N Engl J Med 2006;355:2408-17. [Crossref] [PubMed]
- Holyoake T, Jiang X, Eaves C, et al. Isolation of a highly quiescent subpopulation of primitive leukemic cells in chronic myeloid leukemia. Blood 1999;94:2056-64. [PubMed]
- Savona M, Talpaz M. Getting to the stem of chronic myeloid leukaemia. Nat Rev Cancer 2008;8:341-50. [Crossref] [PubMed]
- Chu S, McDonald T, Lin A, et al. Persistence of leukemia stem cells in chronic myelogenous leukemia patients in prolonged remission with imatinib treatment. Blood 2011;118:5565-72. [Crossref] [PubMed]
- Corbin AS, Agarwal A, Loriaux M, et al. Human chronic myeloid leukemia stem cells are insensitive to imatinib despite inhibition of BCR-ABL activity. J Clin Invest 2011;121:396-409. [Crossref] [PubMed]
- Staal FJ, Chhatta A, Mikkers H. Caught in a Wnt storm: Complexities of Wnt signaling in hematopoiesis. Exp Hematol 2016;44:451-7. [Crossref] [PubMed]
- Staal FJ, Famili F, Garcia Perez L, et al. Aberrant Wnt Signaling in Leukemia. Cancers (Basel) 2016;8. [Crossref] [PubMed]
- Luis TC, Naber BA, Roozen PP, et al. Canonical wnt signaling regulates hematopoiesis in a dosage-dependent fashion. Cell Stem Cell 2011;9:345-56. [Crossref] [PubMed]
- Zhao C, Blum J, Chen A, et al. Loss of beta-catenin impairs the renewal of normal and CML stem cells in vivo. Cancer Cell 2007;12:528-41. [Crossref] [PubMed]
- Heidel FH, Bullinger L, Feng Z, et al. Genetic and pharmacologic inhibition of beta-catenin targets imatinib-resistant leukemia stem cells in CML. Cell Stem Cell 2012;10:412-24. [Crossref] [PubMed]
- Hu Y, Chen Y, Douglas L, et al. beta-Catenin is essential for survival of leukemic stem cells insensitive to kinase inhibition in mice with BCR-ABL-induced chronic myeloid leukemia. Leukemia 2009;23:109-16. [Crossref] [PubMed]
- Neviani P, Harb JG, Oaks JJ, et al. PP2A-activating drugs selectively eradicate TKI-resistant chronic myeloid leukemic stem cells. J Clin Invest 2013;123:4144-57. [Crossref] [PubMed]
- Scheller M, Schönheit J, Zimmermann K, et al. Cross talk between Wnt/beta-catenin and Irf8 in leukemia progression and drug resistance. J Exp Med 2013;210:2239-56. [Crossref] [PubMed]
- Castellone MD, Teramoto H, Williams BO, et al. Prostaglandin E2 promotes colon cancer cell growth through a Gs-axin-beta-catenin signaling axis. Science 2005;310:1504-10. [Crossref] [PubMed]
- Gregory MA, Phang TL, Neviani P, et al. Wnt/Ca2+/NFAT signaling maintains survival of Ph+ leukemia cells upon inhibition of Bcr-Abl. Cancer Cell 2010;18:74-87. [Crossref] [PubMed]
- Grumolato L, Liu G, Haremaki T, et al. beta-Catenin-independent activation of TCF1/LEF1 in human hematopoietic tumor cells through interaction with ATF2 transcription factors. PLoS Genet 2013;9:e1003603. [Crossref] [PubMed]
- Yu S, Li F, Xing S, et al. Hematopoietic and Leukemic Stem Cells Have Distinct Dependence on Tcf1 and Lef1 Transcription Factors. J Biol Chem 2016;291:11148-60. [Crossref] [PubMed]
- Li F, He B, Ma X, et al. Prostaglandin E1 and Its Analog Misoprostol Inhibit Human CML Stem Cell Self-Renewal via EP4 Receptor Activation and Repression of AP-1. Cell Stem Cell 2017;21:359-73.e5. [Crossref] [PubMed]
- Lamb J. The Connectivity Map: a new tool for biomedical research. Nat Rev Cancer 2007;7:54-60. [Crossref] [PubMed]
Cite this article as: Masamoto Y, Kurokawa M. Targeting chronic myeloid leukemia stem cells: can transcriptional program be a druggable target for cancers? Stem Cell Investig 2018;5:10.


