
Interplay between BMP2 and Notch signaling in endothelial-mesenchymal transition: implications for cardiac fibrosis
Introduction
The endothelial-to-mesenchymal transition (EndoMT) is one of the main events occurring in the heart during the development of the organ and its disorders. During EndoMT, endothelial cells undergo several changes, including the loss of tight intercellular junctions, an increased migration activity, and an intensified secretion of extracellular matrix (ECM) proteins. As a result, the cells acquire mesenchymal phenotype (1). For example, this cell transformation is necessary for the correct formation of heart valves in the developing heart during normal physiological development (2). On the contrary, the reactivation of EndoMT is also characteristic of some cardiovascular pathologies, such as atherosclerosis and myocardial fibrosis (3,4).
Many signaling pathways are involved in EndoMT and actively modulate the process, and one of the most important is the Notch signaling, which is an essential regulator of EndoMT in myocardial fibrosis, stimulating the expression of Snai, Slug, and Zeb1 transcription factors (4-6). Notch signaling is an evolutionary conserved cell-to-cell communication pathway involved in cell fate selection through ligand-receptor interactions between transmembrane molecules of neighboring cells observed in embryonic development, angiogenesis, and wound healing (7).
Cardiac fibrosis can develop as a consequence of heart failure, such as in the case of myocardial infarction (MI), and is characterized by excessive collagen deposition, which causes organ remodeling (8,9). Previously, we reported that bone morphogenetic protein 2 (BMP2) factor from the tumor necrosis factor β (TGF-β) superfamily is involved in the processes of early myocardial remodeling in cardiac mesenchymal cells (CMCs) in rats (10). Activation of BMP2 in the CMCs was accompanied by an increase in the expression of Notch signaling components, which indicated the reactivation of embryonic signaling pathways in myocardial fibrosis (11). The signaling cascade induced by TGF-β is the most crucial regulatory pathway in the initiation and development of EndoMT (12). The interplay between the Notch pathway and BMP2 in regulating cardiac fibrosis is complex and needs to be fully understood.
Previously, the group of José Luis de la Pompa (13,14) showed an interaction between Notch pathway and BMP2 in the formation of the cardiac valve in mice through a Snail1-dependent mechanism as a common tissue-specific target of these signaling pathways. In the developing heart, they are antagonistic to each other, and the correct regulation of the expression of target genes ensures the proper formation of the organ due to the specific developmental patterning (15). The Notch signaling inhibits fibrosis in rat MI models by suppressing the transformation of cardiac fibroblasts into myofibroblasts by antagonizing TGF-β1/Smad3 signaling (16), a key regulator of EndoMT. The balance between Notch and BMP2 signaling determines the fate of fibroblasts and the progression of EndoMT and fibrosis in the heart.
We propose that the specific contribution of CMCs to the development of cardiac fibrosis, in the context of EndoMT regulation, is mediated by the interplay between the BMP2 and Notch signaling pathways. Activation of these pathways in CMCs leads to the regulation of common downstream targets, resulting in the modulation of EndoMT markers and subsequent fibrotic processes in the heart.
In this study, we have observed potential evidence suggesting a relationship between Notch signaling and BMP2 in the regulation of EndoMT through a potential SLUG-dependent mechanism (SNAI2) in human CMCs. Co-cultivation of CMCs and HUVECs in two different combinations resulted in opposite cellular responses regarding target gene expression and α-smooth muscle actin (α-SMA) synthesis. Our findings suggest that the activation of BMP2 and Notch is tissue-specific and highly dependent on the cell type being utilized.
Methods
Cell cultures
This study was approved by the local ethics committee of the Almazov National Medical Research Centre (ethical permit 12.26/2014) and complied with the principle of the Declaration of Helsinki (as revised in 2013) (17). All patient representatives gave informed consent.
The following cell types were used in the study:
- CMCs isolated from tissue samples obtained from donors (n=6).
- Human umbilical vein endothelial cells (HUVECs) provided by the cell biobank of Almazov National Medical Research Centre (n=6).
Cultivation of CMCs
CMCs from myocardial tissue were isolated and expanded as described earlier in the studies (18,19). Myocardial tissue was mechanically grinded into small fragments and digested by enzymatic tissue dissociation with collagenase II solution (Worthington, Lakewood, USA) (2 mg/mL) for 1.5 hours at 37 °C. Then the cell suspension was passed through a 40 µm strainer, centrifuged at 300 ×g for 5 min, and the cells were resuspended in culture medium [70% DMEM-F12 (Invitrogen, Thermo Fisher Scientific, New York, USA), 20% ECM (Invitrogen) with supplements, 10% fetal bovine serum (FBS) (HyClone, Cytiva, Logan, USA), 100 µM MEM-NEAA amino acid solution (Gibco, Thermo Fisher Scientific), 2 mM L-glutamine (Gibco), a mixture of penicillin (100 U/mL) and streptomycin (100 µg/mL) (Gibco)], seeded in a flask with a 0.1% gelatin coating and expanded. After expansion in a 25-cm2 flask, the cells were subcultured (passage 1) and kept in Petri dishes (Corning, Tewksbury, USA) in a nutrient medium at 37 °C and 5% CO2. The culture medium was changed every 3 days. For each patient, one CMC line representing a mixed population derived from myocardial tissue was analyzed. All lines were analyzed at the third passage.
Phenotypic characterization of CMC lines was carried out according to the protocol described earlier (20). Cells were resuspended in 100 µL of phosphate-buffered saline (PBS) carrying 1% bovine serum albumin (BSA) (Sigma-Aldrich, St. Louis, MO, USA), incubated for 20 min at 20 °C in the dark with the following monoclonal antibodies: CD31 PE (IM2409, Beckman Coulter, Marseille, France), anti-CD34 APC (IM2472U, Beckman Coulter), anti-CD90 PE (IM1840U, Beckman Coulter), CD146 phycoerythrin (PE) (A07483, Beckman Coulter), anti-CD166 PE (A22361, Beckman Coulter), anti-PDGFRβ allophycocyanin (APC) (FAB1263A, BD Pharmingen, San Jose, USA). Unstained cells were used as a negative control. CMC immunophenotype (data not shown) was assessed by flow cytometric analysis performed on a CytoFlex (Beckman Coulter). The Kaluza 2.0 software (Beckman Coulter) was used for data analysis.
Cultivation of HUVECs
HUVECs were harvested from the umbilical vein by enzymatic dissociation as described (21). Cells were cultured in Petri dishes coated with 0.2% gelatin in a special medium for culturing ECM endothelial cells (ScienCell, Carlsbad, USA).
Co-culture of CMC and HUVEC cell cultures
We co-cultivated CMC and HUVEC cell cultures in two combinations: when CMCs was seeded first, and then HUVECs, and vice versa. The second cell cultures were seeded 24 hours after the first cultures. Cells were cultured in 12-well plates (to isolate total RNA, 75,000 cells per well) and 6-well plates (for α-SMA staining, 25,000 cells per well) coated with 0.2% gelatin in a special medium for culturing ECM endothelial cells (ScienCell).
Production of viral particles
Lentiviral packaging plasmids were a generous gift from D. Trono (École Polytechnique Fédérale de Lausanne, Switzerland); pLVTHM has been modified to carry the intracellular domain of NOTCH1 (NICD) (5). Lentiviral particles were obtained using the packaging line HEK293-T, as described (22). HEK293-T cells were cultured in DMEM medium (Gibco) supplemented with 10% FBS (Gibco) and 1% antibiotic penicillin-streptomycin (Invitrogen) in an incubator (37 °C, 5% CO2, humidity 99%). The efficiency of transgene expression with NICD virus was confirmed by immunoblotting with anti-NOTCH1 antibodies (SC6014, Santa Cruz, Dallas, USA) and quantitative polymerase chain reaction (qPCR) with primers for the Notch HEY1 target gene (data not shown).
Transduction of cell cultures with lentiviral vectors NICD gene and BMP2 gene
Lentivirus transduction was performed during cell culture seeding by adding 25 µL of viral concentrate carrying either NICD or BMP2. In the case of double transduction, the second NICD vector was introduced 18 hours after cell seeding and the initial transduction with the lentivirus carrying BMP2.
Immunocytochemical staining
The cell cultures, 14 days after seeding, were fixed on slides with 4% paraformaldehyde for 12 minutes on ice. After removing the fixative, the cells were washed three times for 5 minutes with PBS and treated with 0.5% Triton X-100 in PBS for 3 minutes. Following three additional washes with PBS, the cells were incubated with 1% BSA in PBS for 40 minutes. The coverslip area was then marked with a hydrophobic marker, and primary antibodies against ACTA-2 (Novocastra, Wetzlar, Germany) were applied at a 1:100 dilution in BSA. The cells were incubated in a humidity chamber at room temperature for 1 hour, followed by three 5-minute washes with BSA. Secondary antibodies conjugated with Alexa Fluor 647, diluted 1:1,000, were applied and incubated for 40 minutes in the dark at room temperature. After three additional 5-minute washes with BSA, the nuclei were stained with 4',6-diamidino-2-phenylindole (DAPI) (Invitrogen, USA) at a dilution of 1:10,000 in BSA for 5 seconds. Finally, coverslips with cells were mounted onto slides using Fluoromount (Sigma, USA). The preparations were examined using an Axio Observer fluorescence microscope (Carl Zeiss, Oberkochen, Germany), and the images were processed with the Axiovision 4.7 software (Carl Zeiss).
Real-time PCR (RT-PCR) reaction
Total RNA was extracted from CMCs and HUVECs using Extract RNA reagent (Eurogen, Moscow, Russia) according to the manufacturer’s instructions. All samples were pretreated with DNase. Total RNA (1 µg) was reverse transcribed with an MMLV RT Kit (Eurogen). We used a 5-fold reaction mixture qPCR mix-HS SYBR with intercalating dye SYBR Green I (Eurogen) for RT-PCR. Complementary DNA (cDNA) (50 ng), forward and reverse primers (10 µM each) were added to the mixture, and the final volume was adjusted with sterile water to 25 µL. The sequences of the primers used were as follows:
- GAPDH (F: 5'-ACAACTTTGGTATCGTGGAAGG-3', R: 5'-CAGTAGAGGCAGGGATGATGTT-3');
- NOTCH1 (F: 5'-GAGGCGTGGCAGACTATGC-3', R: 5'-CTTGTACTCCGTCAGCGTGA-3');
- HEY1 (F: 5'-TGAGCTGAGAAGGCTGGTAC-3', R: 5'-ATCCCAAACTCCGATAGTCC-3');
BMP2 (F: 5'-AGCCAGCCGAGCCAACAC-3', R: 5'-CCCACTCGTTTCTGGTAGTTCTTC-3'); - RUNX2 (F: 5'-GAGTGGACGAGGCAAGAGTT-3', R: 5'-GGGTTCCCGAGGTCCATCTA-3');
- SNAI1 (F: 5'-CTCTTTCCTCGTCAGGAAGC-3', R: 5'-GGCTGCTGGAAGGTAAACTC-3');
- SNAI2 (F: 5'-TCCAGACCCTGGTTGCTTCA-3', R: 5'-GAATGGGTCTGCAGATGAGCC-3').
RT-PCR was performed on a LightCycler480 system (Roche Diagnostics, Indianapolis, USA) using specific forward and reverse primers for target genes. qPCR was performed for 45 cycles. Data analysis was conducted using the 2−ΔΔCt method; relative gene expression was normalized on the GAPDH housekeeping gene.
qPCR data on gene expression were analyzed using GraphPad Prism 9.4.1 (458) (GraphPad Software, San Diego, CA, USA). Values are expressed as median and interquartile range. Groups were compared using the unpaired nonparametric Mann-Whitney test. The P≤0.05 value was considered significant.
Results
Activation of Notch and BMP2 signaling pathways modulates target gene expression in CMCs and HUVECs
In order to evaluate the effect of activating Notch signaling and BMP2 on the expression of target genes, we transduced CMCs and HUVECs with a lentiviral vector carrying either the NICD (active form of NOTCH1) or BMP2 sequences. After 48 hours, we isolated RNA for RT-PCR. An empty vector [transduction control (TRC)] that does not carry insert genes was used as a control to evaluate the effect of cell transduction, and the cells without any induction were also included.
We observed that the activation of NICD resulted in an elevated expression of the Notch target gene HEY1 and the NOTCH1 receptor in both cell cultures (Figure 1), indicating successful cell transduction with the NICD lentiviral vector. However, only HUVECs showed an increase in BMP2 expression in response to the introduction of the NICD vector, while there was no change in BMP2 expression in CMCs. Furthermore, RUNX2 was suppressed in the CMCs.
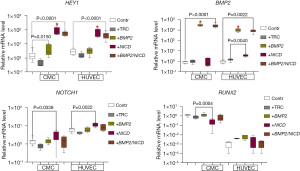
Upon introducing the BMP2 lentiviral vector into the cell cultures, we observed an increased expression of BMP2 in both cell cultures, confirming the effectiveness of this vector (Figure 1). Additionally, the induction of BMP2 led to the activation of HEY1 in the CMCs.
To investigate the potential interplay between Notch and BMP2 signaling pathways, we co-transduced CMC and HUVEC cell cultures with lentiviral vectors carrying NICD and BMP2 sequences, respectively. The BMP2 vector was introduced first, followed by NICD 18 hours later (+BMP2/NICD, Figure 1). Our results showed that co-transduction did not enhance target gene expression compared to a single pathway induction.
Notch and BMP2 activation induces increased α-SMA synthesis, except in HUVECs in response to BMP2 gene induction
In order to evaluate the effect of Notch signaling or BMP2 activation on the expression of EndoMT markers, we transduced CMCs and HUVECs with a lentiviral vector carrying the NICD or BMP2 sequences, respectively. Upon Notch induction (Figure 2A), we observed active transcription of SNAI2 (SLUG) in both types of cells, in contrast to SNAI1, which was overexpressed only in response to BMP2 induction in CMCs. Furthermore, the expression of SNAI2 in HUVECs increased to a level comparable to the baseline expression observed in control mesenchymal cells. However, double transduction of CMCs and HUVECs by BMP2 and NICD did not increase the expression level of EndoMT marker genes (+BMP2/NICD, Figure 2A).
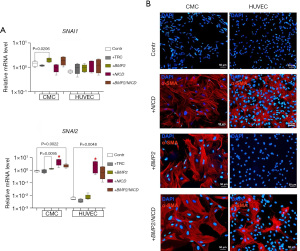
α-SMA is a reliable indicator of the EndoMT in endothelial cells or when mesenchymal cells are activated during development or injury (23-26). To assess the activation of EndoMT in CMC and HUVEC cultures in response to Notch and BMP2 signaling induction, we performed immunostaining for α-SMA. We observed (Figure 2B) that CMCs, like HUVECs, did not synthesize α-SMA in control without induction. Notch activation led to α-SMA accumulation in both CMCs and HUVECs (Figure 2B). However, BMP2 transduction of cell cultures contributed to the synthesis of α-SMA only in CMCs, which is in accordance to accumulation of SNAI1 and SNAI2 transcripts by qPCR. As a result of the introduction of both vectors, we observed EndoMT activation in both CMCs and HUVECs (Figure 2B). However, the synthesis of α-SMA was less pronounced in HUVECs than in CMCs.
Co-cultivation of CMCs and HUVECs alters the expression of target genes, with specific activation of HEY1 gene and BMP2 gene in double transduction of CMC/HUVEC cultures
Given that Notch signaling is known to mediate intercellular communication, we aimed to investigate the impact of co-culturing mesenchymal and endothelial cells on the expression of the Notch and BMP2 pathways.
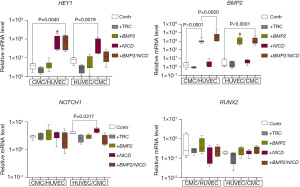
Upon introducing the NICD lentiviral vector, we successfully observed an upregulation of HEY1 expression in both co-cultivations. However, the expression of NOTCH1 increased only in the HUVEC/CMC co-culture. Similarly, transducing the cell cultures with the BMP2 vector resulted in the activation of BMP2 in both co-cultures, affirming the effectiveness of the vectors when used in a co-cultured environment.
Our findings (Figure 3) reveal the expression profile variability in the cell cultures, which appears to be influenced by the order of cell seeding. Specifically, a decrease in the expression of target genes was observed compared to monoculture conditions. Nevertheless, we noted a stimulating effect on the expression of HEY1 and BMP2 in response to double transduction, particularly in the combination of CMC/HUVEC cultures (+BMP2/NICD, Figure 3).
Induction of α-SMA synthesis in CMCs is modulated by BMP2 and BMP2/NICD activation during co-cultivation, and its correlation with SNAI2 expression
To evaluate the effect of co-cultivating CMCs and HUVECs on α-SMA synthesis in response to the induction of Notch and BMP2 signaling pathways, we immunostained cell co-cultures for α-SMA. Our results (Figure 4) suggest that activation of the Notch signaling induces α-SMA synthesis in both CMC and HUVEC co-culture, regardless of the seeding order. Double transduction of the CMC + HUVEC co-culture resulted in an increase in BMP2 expression (+BMP2/NICD, Figure 3) and, conversely, a suppression of the expression of EndoMT markers (+BMP2/NICD, Figure 4A). This was reflected in the suppression of α-SMA accumulation of in the presence of a single BMP2 vector, as well as of both BMP2 and NICD vectors (+BMP2/NICD, Figure 4B).
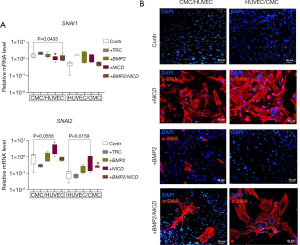
BMP2 transduction of cell cultures had an inhibitory effect on EndoMT in mesenchymal cells, and was seen primarily by SNAI2 expression decrease, when grown together with endothelial cells. Activation of both Notch and BMP2 pathways led to the induction of EndoMT in HUVEC/CMC co-cultures (+BMP2/NICD, Figure 4B), and this correlated with SNAI2 accumulation (+BMP2/NICD, Figure 4A). In contrast, we observed that the expression of SNAI1 was suppressed in response to Notch induction and double transduction in the CMC/HUVEC co-culture.
Discussion
EndoMT is a critical process involved in cardiovascular system development and disorders (27). While embryonic signaling pathways like Notch and Bmp have been shown to regulate EndoMT, their interactions in this process are not yet fully understood. Consequently, identifying a way to regulate these pathways is a priority for treating cardiovascular pathologies in adulthood.
To investigate this, we induced Notch and Bmp signaling pathways (specifically BMP2) in CMCs and HUVECs using various combinations. We transduced the cells with vectors carrying NICD and/or BMP2 and isolated RNA 48 hours later for RT-PCR to evaluate the expression of Notch and Bmp pathway components and activation of EndoMT markers. We used α-SMA immunostaining to indicate EndoMT 14 days after plating.
Notch signaling is a highly conserved juxtracrine signaling pathway that includes receptors (like NOTCH1), ligands, and target genes (like HEY1). In the canonical Notch signaling, the receptor and ligand of the two neighboring cells interact, leading to proteolytic cleavage of the receptor’s intracellular part and translocation of NICD into the nucleus to activate target gene expression. We induced Notch signaling in CMCs and HUVECs using a lentiviral vector carrying the NICD sequence. After activation, both cultures showed an increase in HEY1 and NOTCH1 receptor expression. Furthermore, the transduction of NICD cell cultures led to the synthesis and subsequent accumulation of α-SMA in both CMCs and HUVECs after 14 days of induction. We found that Notch pathway activation using NICD transduction increased BMP2 expression in HUVECs but had no effect on CMCs. We observed a decrease in RUNX2 expression, one of the Bmp target genes (28,29), in CMCs, indicating two opposite processes in endothelial and mesenchymal cells.
BMP2 is a growth factor and a component of the TGF-β superfamily that plays a role in many developmental processes, including cardiogenesis, neurogenesis, and osteogenesis (30-32). BMP2 has also been noted in epithelial-to-mesenchymal transition (EMT) processes in development (33,34). Notch signaling has been shown to regulate BMP2 during osteogenic differentiation (35). However, its precise regulation in the context of EndoMT and its interactions with other embryonic signaling pathways, including Notch, are still not fully understood. Transduction of BMP2 cell cultures successfully upregulated BMP2 expression in CMCs and HUVECs. We found that induction of BMP2 in CMCs resulted in an increase in the expression of HEY1 (the Notch target gene) and had no effect on the change in NOTCH1, which may indicate interaction only with downstream targets. At the same time, activation of BMP2 in HUVECs had no effect on the expression of Notch signaling pathway components. Moreover, the introduction of BMP2 through transduction had an inducing effect specifically on CMCs, resulting in the synthesis and accumulation of α-SMA. However, in contrast to CMCs, HUVECs did not respond to BMP2 activation, suggesting a potential tissue-specific effect of this signaling pathway.
During cardiac EMT, endocardial cells undergo significant changes in gene expression, including Notch-dependent induction of ACTA2 (α-SMA), SNAI1, and SNAI2 (5). Despite differences in BMP2 activation (Figure 1), we observed an increase in SNAI2 expression in both cultures after NICD introduction (Figure 2A). However, this did not affect the expression of SNAI1 (Figure 2A).
BMP2 transduction led to an increase in BMP2 expression in both cultures, and the expression of EndoMT markers differed between CMCs and HUVECs. While SNAI2 was activated in both cultures, only CMCs expressed SNAI1. The coordinated activity of Notch and BMP2 signaling has been noted previously in the developmental processes of the cardiovascular system (13). Ectopic expression of one often led to suppression of the other signaling system in the same tissue. However, coordinated signaling has also been described, for example, between the myocardium and endocardium, as well as during the formation of the heart valve rudiment with SNAI1 activation and influence on EMT processes (36).
We alternately activated the Notch and Bmp signaling pathways in CMCs and HUVECs by introducing the BMP2 vector. Then a second vector bearing NICD was added. We observed that double transduction did not increase the expression of signaling pathway markers and EndoMT in both cultures compared to single induction, and most likely reduced the efficiency of their expression. Interestingly, when examining the impact of double transduction, we did not observe a significant effect on the accumulation of α-SMA in CMCs. The levels of α-SMA were comparable to those observed with single induction, indicating that double transduction also led to the synthesis of α-SMA. However, the effect was less pronounced in HUVECs, indicating that these signaling pathways may have opposing effects in this particular cell type.
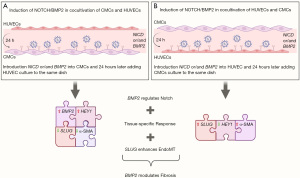
We hypothesized that the interaction between Notch and BMP2 signaling might change when mesenchymal and endothelial cells communicate. CMCs and HUVECs were sown alternately with a difference of 24 hours in two combinations. Evaluation of Notch, BMP2, and EndoMT gene expression in response to induction showed a decrease in the expression of target genes compared to monoculture. Despite using the same cell cultures in various co-cultivation combinations, we observed distinct responses to the introduction of NICD and BMP2 in terms of HEY1 and NOTCH1 expression. In the CMCs/HUVECs combination, we observed a consistent increase in HEY1 expression upon the addition of NICD and BMP2/NICD. However, in the HUVECs/CMCs combination, only the introduction of the NICD vector led to HEY1 activation. The observed discrepancy implies a possible impact of the seeding order on intercellular communication, potentially influenced by the initial colonization predominance of HUVECs in the culture dish. Furthermore, we noted NOTCH1 activation in the HUVEC/CMC seeding combination in response to NOTCH induction, reinforcing our observation of tissue specificity. These findings suggest a potential influence of cell type interaction and seeding order on the modulation of marker expression and signaling pathways.
We observed a decrease in the expression of EndoMT markers such as SNAI1 and SNAI2 in CMC/HUVEC co-culture compared to CMC monoculture in response to BMP2 induction, which was confirmed by the absence of α-SMA synthesis in immunocytochemical staining. This could mean that the addition of the HUVEC culture, which previously did not synthesize α-SMA in monoculture, prevented the manifestation of the indicator in the susceptible CMC culture. At the same time, double transduction led to a significant increase in BMP2 expression in the CMC/HUVEC co-culture compared to single induction, which may indicate a complementary effect in the presence of both factors in the co-culture. In contrast, in HUVEC/CMC co-culture, double transduction positively influenced the accumulation of α-SMA and increased SNAI2 expression. During the experiment, we repeatedly observed that SNAI2 expression directly correlated with the synthesis of α-SMA in cultures in response to either Notch or BMP2 activation.
BMP2, like Notch, plays an essential role in altering the expression of EndoMT marker genes and components of the Notch and Bmp signaling pathways, and appears to promote Notch deregulation possibly through a Slug-dependent mechanism (SNAI2). However, our results showed that in both CMCs and HUVECs, the relationship in intercellular communications are changed during co-cultivation, leading to the formation of antagonistic effects in gene expression, which are comparable when choosing a cell fate between developing tissues.
The opposite cellular response observed in the co-cultivation of CMCs and HUVECs in different combinations can be attributed to the complex interplay between the cell types and the signaling mechanisms involved (Figure 5). In one study, where endothelial cells were co-cultured with mesenchymal stem cells (MSCs), the endothelial cells were found to induce smooth muscle cell (SMC) differentiation in the MSCs (37). This induction of SMC fate was mediated through Notch signaling and was independent of growth factor signaling. On the other hand, where endothelial cells were co-cultured with bone marrow stem cells (BMSCs), both osteogenesis and angiogenesis were induced (38). The co-cultured cells exhibited enhanced expression of osteogenic genes, suggesting a supportive role of endothelial cells in promoting both bone formation and blood vessel formation. These findings suggest that the specific combination of cell types and their microenvironment can influence the cellular response and fate determination, highlighting the importance of cell-cell interactions and the context in which they occur.
While we have observed signs of EndoMT in cell cultures, by evaluating the expression of several marker genes and staining for α-SMA, we have not considered a wider range of markers that could expand our work and our panel of genes, namely endothelial markers such as vascular endothelial (VE)-cadherin, endothelial nitric oxide synthase (eNOS) and/or CD31; and mesenchymal-specific markers such as N-cadherin, FSP-1, vimentin and/or fibronectin.
Conclusions
In conclusion, the interaction between the Notch pathway and BMP2 in the context of EndoMT is crucial in regulating cardiac fibrosis. The balance between Notch and BMP2 signaling determines the outcome of EndoMT and fibrosis in the heart. Further research is needed to better understand the mechanisms underlying the interaction between Notch and BMP2 in the heart and to develop new therapeutic strategies for treating cardiac fibrosis.Click or tap here to enter text.
Acknowledgments
We express our deep gratitude to Dr. Ivan Kozyrev, surgeon in Department of Pediatric Cardiac Surgery, Almazov National Medical Research Centre, for his help in obtaining surgical material for cell isolation.
Funding: This study was funded by the Republican Research Scientific and Consulting Center of Expertise (grant No. 4664.2022.1.4).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://sci.amegroups.com/article/view/10.21037/sci-2023-019/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013) and approved by the local ethics committee of the Almazov National Medical Research Centre (ethical permit 12.26/2014). All patient representatives gave informed consent.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Kovacic JC, Mercader N, Torres M, et al. Epithelial-to-mesenchymal and endothelial-to-mesenchymal transition: from cardiovascular development to disease. Circulation 2012;125:1795-808. [Crossref] [PubMed]
- Bischoff J. Endothelial-to-Mesenchymal Transition. Circ Res 2019;124:1163-5. [Crossref] [PubMed]
- Zhang J, Ogbu SC, Musich PR, et al. The Contribution of Endothelial-Mesenchymal Transition to Atherosclerosis. Int J Transl Med 2021;1:39-54. [Crossref]
- Cheng W, Li X, Liu D, et al. Endothelial-to-Mesenchymal Transition: Role in Cardiac Fibrosis. J Cardiovasc Pharmacol Ther 2021;26:3-11. [Crossref] [PubMed]
- Kostina AS, Uspensky VE, Irtyuga OB, et al. Notch-dependent EMT is attenuated in patients with aortic aneurysm and bicuspid aortic valve. Biochim Biophys Acta 2016;1862:733-40. [Crossref] [PubMed]
- Kachanova O, Lobov A, Malashicheva A. The Role of the Notch Signaling Pathway in Recovery of Cardiac Function after Myocardial Infarction. Int J Mol Sci 2022;23:12509. [Crossref] [PubMed]
- Bocci F, Onuchic JN, Jolly MK. Understanding the Principles of Pattern Formation Driven by Notch Signaling by Integrating Experiments and Theoretical Models. Front Physiol 2020;11:929. [Crossref] [PubMed]
- Mocumbi AO, Stothard JR, Correia-de-Sá P, et al. Endomyocardial Fibrosis: an Update After 70 Years. Curr Cardiol Rep 2019;21:148. [Crossref] [PubMed]
- Frangogiannis NG. Cardiac fibrosis: Cell biological mechanisms, molecular pathways and therapeutic opportunities. Mol Aspects Med 2019;65:70-99. [Crossref] [PubMed]
- Docshin PM, Karpov AA, Mametov MV, et al. Mechanisms of Regenerative Potential Activation in Cardiac Mesenchymal Cells. Biomedicines 2022;10:1283. [Crossref] [PubMed]
- He X, Du T, Long T, et al. Signaling cascades in the failing heart and emerging therapeutic strategies. Signal Transduct Target Ther 2022;7:134. [Crossref] [PubMed]
- Mercado-Pimentel ME, Runyan RB. Multiple transforming growth factor-beta isoforms and receptors function during epithelial-mesenchymal cell transformation in the embryonic heart. Cells Tissues Organs 2007;185:146-56. [Crossref] [PubMed]
- Luna-Zurita L, Prados B, Grego-Bessa J, et al. Integration of a Notch-dependent mesenchymal gene program and Bmp2-driven cell invasiveness regulates murine cardiac valve formation. J Clin Invest 2010;120:3493-507. [Crossref] [PubMed]
- MacGrogan D, Luna-Zurita L, de la Pompa JL. Notch signaling in cardiac valve development and disease. Birth Defects Res A Clin Mol Teratol 2011;91:449-59. [Crossref] [PubMed]
- Rutenberg JB, Fischer A, Jia H, et al. Developmental patterning of the cardiac atrioventricular canal by Notch and Hairy-related transcription factors. Development 2006;133:4381-90. [Crossref] [PubMed]
- Zhou XL, Fang YH, Wan L, et al. Notch signaling inhibits cardiac fibroblast to myofibroblast transformation by antagonizing TGF-β1/Smad3 signaling. J Cell Physiol 2019;234:8834-45. [Crossref] [PubMed]
- World Medical Association Declaration of Helsinki. ethical principles for medical research involving human subjects. JAMA 2013;310:2191-4. [Crossref] [PubMed]
- Smits AM, van Vliet P, Metz CH, et al. Human cardiomyocyte progenitor cells differentiate into functional mature cardiomyocytes: an in vitro model for studying human cardiac physiology and pathophysiology. Nat Protoc 2009;4:232-43. [Crossref] [PubMed]
- Kozyrev I, Dokshin P, Kostina A, et al. Dysregulation of Notch signaling in cardiac mesenchymal cells of patients with tetralogy of Fallot. Pediatr Res 2020;88:38-47. [Crossref] [PubMed]
- Kozyrev IA, Golovkin AS, Ignatieva ES, et al. Characterization of mesenchymal heart cells obtained from patients with tetralogy of Fallot and ventricular septal defect. Translational Medicine 2019;6:16-23. [Crossref]
- Kostina A, Shishkova A, Ignatieva E, et al. Different Notch signaling in cells from calcified bicuspid and tricuspid aortic valves. J Mol Cell Cardiol 2018;114:211-9. [Crossref] [PubMed]
- Malashicheva A, Kanzler B, Tolkunova E, et al. Lentivirus as a tool for lineage-specific gene manipulations. Genesis 2007;45:456-9. Erratum in: Genesis 2007;45:793. [Crossref] [PubMed]
- Shinde AV, Humeres C, Frangogiannis NG. The role of α-smooth muscle actin in fibroblast-mediated matrix contraction and remodeling. Biochim Biophys Acta Mol Basis Dis 2017;1863:298-309. [Crossref] [PubMed]
- Nakajima Y, Mironov V, Yamagishi T, et al. Expression of smooth muscle alpha-actin in mesenchymal cells during formation of avian endocardial cushion tissue: a role for transforming growth factor beta3. Dev Dyn 1997;209:296-309. [Crossref] [PubMed]
- Piera-Velazquez S, Li Z, Jimenez SA. Role of endothelial-mesenchymal transition (EndoMT) in the pathogenesis of fibrotic disorders. Am J Pathol 2011;179:1074-80. [Crossref] [PubMed]
- Kovacic JC, Dimmeler S, Harvey RP, et al. Endothelial to Mesenchymal Transition in Cardiovascular Disease: JACC State-of-the-Art Review. J Am Coll Cardiol 2019;73:190-209. [Crossref] [PubMed]
- Anbara T, Sharifi M, Aboutaleb N. Endothelial to Mesenchymal Transition in the Cardiogenesis and Cardiovascular Diseases. Curr Cardiol Rev 2020;16:306-14. [Crossref] [PubMed]
- Zamurovic N, Cappellen D, Rohner D, et al. Coordinated activation of notch, Wnt, and transforming growth factor-beta signaling pathways in bone morphogenic protein 2-induced osteogenesis. Notch target gene Hey1 inhibits mineralization and Runx2 transcriptional activity. J Biol Chem 2004;279:37704-15. [Crossref] [PubMed]
- Sharff KA, Song WX, Luo X, et al. Hey1 basic helix-loop-helix protein plays an important role in mediating BMP9-induced osteogenic differentiation of mesenchymal progenitor cells. J Biol Chem 2009;284:649-59. [Crossref] [PubMed]
- Lavery K, Swain P, Falb D, et al. BMP-2/4 and BMP-6/7 differentially utilize cell surface receptors to induce osteoblastic differentiation of human bone marrow-derived mesenchymal stem cells. J Biol Chem 2008;283:20948-58. [Crossref] [PubMed]
- Seeherman HJ, Berasi SP, Brown CT, et al. A BMP/activin A chimera is superior to native BMPs and induces bone repair in nonhuman primates when delivered in a composite matrix. Sci Transl Med 2019;11:eaar4953. [Crossref] [PubMed]
- Tanaka K, Kaji H, Yamaguchi T, et al. Involvement of the osteoinductive factors, Tmem119 and BMP-2, and the ER stress response PERK-eIF2α-ATF4 pathway in the commitment of myoblastic into osteoblastic cells. Calcif Tissue Int 2014;94:454-64. [Crossref] [PubMed]
- Gorelova A, Berman M, Al Ghouleh I. Endothelial-to-Mesenchymal Transition in Pulmonary Arterial Hypertension. Antioxid Redox Signal 2021;34:891-914. [Crossref] [PubMed]
- Ma L, Lu MF, Schwartz RJ, et al. Bmp2 is essential for cardiac cushion epithelial-mesenchymal transition and myocardial patterning. Development 2005;132:5601-11. [Crossref] [PubMed]
- Shimizu T, Tanaka T, Iso T, et al. Notch signaling pathway enhances bone morphogenetic protein 2 (BMP2) responsiveness of Msx2 gene to induce osteogenic differentiation and mineralization of vascular smooth muscle cells. J Biol Chem 2011;286:19138-48. [Crossref] [PubMed]
- Andrés-Delgado L, Galardi-Castilla M, Münch J, et al. Notch and Bmp signaling pathways act coordinately during the formation of the proepicardium. Dev Dyn 2020;249:1455-69. [Crossref] [PubMed]
- Lin CH, Lilly B. Endothelial cells direct mesenchymal stem cells toward a smooth muscle cell fate. Stem Cells Dev 2014;23:2581-90. [Crossref] [PubMed]
- Gurel Pekozer G, Torun Kose G, Hasirci V. Influence of co-culture on osteogenesis and angiogenesis of bone marrow mesenchymal stem cells and aortic endothelial cells. Microvasc Res 2016;108:1-9. [Crossref] [PubMed]
Cite this article as: Docshin P, Bairqdar A, Malashicheva A. Interplay between BMP2 and Notch signaling in endothelial-mesenchymal transition: implications for cardiac fibrosis. Stem Cell Investig 2023;10:18.


