The paradox of Foxd3: how does it function in pluripotency and differentiation of embryonic stem cells?
Understanding the precise regulatory mechanisms that control progenitor cell pluripotency, lineage commitment, and differentiation are fundamental questions in development and stem cell biology. The use of embryonic stem cells (ESCs), reprogramming of induced pluripotent stem cells (iPSCs), and development of novel culture protocols, including serum replacements for ESC culture (1-3), has rapidly increased our understanding of various pluripotent states. Over the past decade, researchers have developed in vitro techniques that recapitulate many of the developmental transitions of the early mammalian embryo and subsequent differentiation to each of the embryonic germ layers (3-5) (Figure 1). ESCs can be derived from the inner cell mass of the blastocyst or epiblast of the pre-implantation embryo and maintained in the “naïve” early epiblast stage when grown in LIF and 2i (MAPK and GSK3 inhibitors) (3) (Figure 1A-D). More recently, in vitro models have also been developed for post-implantation “primed” epiblast progenitors including differentiation of ESCs into 2 intermediate pre-gastrulation states, epiblast cells (EpiCs) and epiblast-like cells (EpiLCs) (4-6) (Figure 1E-G). EpiLCs can be differentiated into primordial germ-like cells (PGCLCs), a process that involves reactivation of a subset of naïve genes and activation of PGC-specific gene expression (5) (Figure 1G,J). In addition to these transient stem cell populations, epiblast stem cells (EpiSCs) can be derived from the epiblast of E5.5–6.5 embryos (or differentiation of ESCs) and are molecularly similar to late gastrulating epiblast progenitors (10,11) (Figure 1H,I). This improved methodology provides researchers with unique temporal access and allows for a more complete understanding of the molecular, transcriptional, and epigenetic processes driving development at each stage.
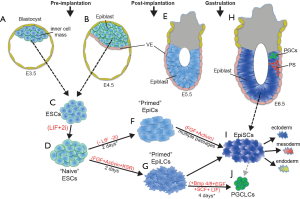
One potential regulatory factor during early to late epiblast transition is the transcription factor Foxd3. Foxd3, a member of the forkhead family, has been shown to play a key role in maintaining the stem cell properties of ESCs, trophoblast stem cells, neural crest progenitor cells, and skin-derived precursor cells (12-18). While the in vivo and in vitro requirements for Foxd3 in these cell lineages have been established, the mechanisms by which Foxd3 elicits its functions is less clear. Two publications from earlier this year dove deeply into understanding how Foxd3 regulates gene expression during early cell fate transitions of progenitor cells (6,7). While the two publications shared some similarity, they also presented some conflicting results.
Two models of Foxd3 function during transitions from “naïve” to “primed” pluripotency
Krishnakumar et al. were interested in understanding the role of Foxd3 in the transition from ESCs to EpiCs, which are molecularly similar to the late epiblast (6) (Figure 1D,F). They found that Foxd3 preferentially binds mutually exclusive enhancer sites in ESCs compared to EpiCs. In ESCs, Foxd3 primarily functions as a repressor, through recruitment of histone deacetylases (HDACs). Relocalization of Foxd3 during the ESC to EpiC transition was associated with nucleosome depletion and H3K4me1 (a mark of active/poised enhancers) at newly bound loci. Furthermore, their work reveals that Foxd3 initiates nucleosome removal and a “primed” state at targets through recruitment of the nucleosome remodeler Brg1, a member of the NuRD complex. Interestingly, while these data suggest a role for Foxd3 in establishment of enhancers for future activation, Foxd3 binding did not correlate with increased H3K27ac (active histone marks) at EpiC binding sites. Instead, the authors found that after nucleosome remodeling, Foxd3 repressed gene expression via HDAC recruitment. The work published by Krishnakumar et al. highlights dual-mechanisms of action: Foxd3 is responsible for opening EpiC loci via Brg1/NuRD remodeling while simultaneously maintaining a repressed state in these enhancers through its interactions with HDACs.
Respuela et al. sought to identify the mechanisms of Foxd3 function during transitions between naïve and primed pluripotency and focused their study on the transition from ESCs to EpiLCs and in the differentiation of EpiLCs to PGCLCs (7) (Figure 1D,G,J). Similar to Krishnakumar et al., they found that Foxd3 largely functions as a repressor and show a requirement for Foxd3 in exit from the naïve state. Their data indicate that the repressive function of Foxd3 is mediated through inactivation of enhancers that are associated with key naïve pluripotency and PGC genes. Further, Respuela et al. propose that Foxd3 regulates local chromatin structure to decommission active enhancers by recruitment of the histone demethylase Lsd1, a member of the NuRD complex, and a reduction of activating factors such as p300 (7). They extend their study to show that Foxd3 expression must be extinguished to allow for reactivation of naïve and PGC genes during specification of PGCs from EpiLCs.
Molding a combined model for the role of Foxd3 during early differentiation of ESCs
On the surface, it appears that the findings of these papers are somewhat contradictory. For example, Respuela et al. found that Foxd3 expression increases as ESCs transition to a primed EpiLC state and that Foxd3 promotes naïve pluripotency exit by binding and inactivating the active enhancers of highly expressed naïve genes. In contrast, Krishnakumar et al. found Foxd3 to be expressed at equivalent levels in ESCs and EpiCs, but was bound to distinct enhancers in each cell type. Foxd3 binding sites in ESCs were adjacent to moderately expressed genes, and expression of these genes increased to maximal levels only after re-localization of Foxd3 to EpiC-specific sites. Seemingly, the largest difference between the manuscripts is the lack of overlap between the published ChIP-Seq data (19). Indeed, a comparison between the data found by both groups indicated only a 12% overlap in Foxd3 peaks. The differences between the ChIP-Seq data from both groups is likely due to a combination of technical differences including different tagging methods and control DNA, computational variation and perhaps most importantly, differences in ESC culture conditions, differentiation protocols and timing of differentiation into what are likely distinct states of primed epiblast progenitor cells. As mentioned by Sweet, this may serve as a cautionary tale for those intending to conduct and interpret next generation sequencing experiments (19).
While the groups asked fundamentally different questions, the papers may be quite complementary, with each dataset providing a snapshot during progression from naïve to primed pluripotency. Both Krishnakumar et al. and Respuela et al. found that Foxd3 primarily functions as a repressor and interacts with NuRD complex members to mediate loss of H3K27ac (6,7,19). While it is difficult to interpret the ChIP-Seq data towards a single model, one interpretation is that examination of Foxd3 binding in ESCs grown in serum + LIF (SL) conditions provides poor resolution of binding during differentiation due to heterogeneity of cells with regard to various states of naïve and primed pluripotency. Not surprisingly, ChIP-Seq from SL-ESCs and EpiLCs did not detect many differences in Foxd3 sites, whereas distinct Foxd3 binding sites were found in the transition from 2i-ESCs to EpiCs.
It is plausible that both publications present an accurate, yet incomplete, picture of the mechanisms by which Foxd3 functions. Krishnakumar et al. discussed the hypothesis that the conserved winged-helix domain of Foxd3 allows it to bind condensed chromatin and nucleosomes and may function as a pioneer factor (6). The model of Foxd3 as a pioneer factor that can establish enhancer accessibility and recruit other factors and histone modifiers to control local chromatin structure is also consistent with the Foxd3-Lsd1/NuRD interaction and enhancer decommissioning reported in Respuela et al. (7). Alternatively, Respuela et al. proposed a model by which Foxd3 is bound to active enhancers, and then, in the context of interactions with co-regulators is required to decommission active enhancers. These decommissioned enhancers are not completely inactivated and remain primed for future activation in differentiating PGCLCs (7). Similarly, Krishnakumar et al. presents data to demonstrate that Foxd3 binding is associated with displacement of nucleosomes. The locus is then actively repressed by virtue of NuRD/HDAC inhibition of H3K27ac but remains primed for future activation (6). Both models suggest that Foxd3 plays an active role in gene repression the earlier progenitor stages while playing a more passive role in later stages.
Another consideration is the direct or indirect role of Foxd3 in differentiated cell lineages. For example, Liu and Labosky demonstrated that, in the absence of Foxd3, mESCs prematurely differentiate into mesendoderm lineages while markers of the ectoderm are reduced (13). Similarly, previous gene expression analysis following loss of Foxd3 in ESCs identified genes required for differentiation to multiple lineages as possible Foxd3 targets (15). Furthermore, Foxd3 is required for distinct cell fate choices in early vs. later neural crest progenitors; in early progenitors Foxd3 is required to repress mesenchymal fates in order to maintain neural fates (14). In contrast, when deleted in later enteric nervous system progenitors, Foxd3 is required for glial but not neural fates (20). These examples highlight the possibility that in distinct progenitors types, and even in the context of distinct stages of pluripotency/differentiation, Foxd3 may have context-dependent co-factor interactions or binding that allow it dynamically change its function.
Summary
What remains clear after reviewing both publications is the fact that Foxd3 is a key regulator of gene expression during progression from naïve to primed pluripotency. Both Krishnakumar et al. and Respuela et al. provide important insights into pluripotency in the early epiblast and have identified several novel roles for Foxd3. It is quite possible that Foxd3 functions include the stage-dependent establishment and active repression of “primed” epiblast enhancers, decommissioning of naïve enhancers, and progressive release of repression at “primed” and/or lineage specific binding sites. Future studies may uncover more mechanisms and co-regulators that allow Foxd3 to scan condensed chromatin, temporally distinguish proper binding sites and appropriately regulate gene expression at its targets. Answers to these questions are likely to lead to better understanding of Foxd3 function, and moreover, are relevant to our understanding of how cell fate transitions occur in stem cell biology and throughout development.
Acknowledgements
JL Plank-Bazinet completed this work as part of her official duties at the National Institutes of Health (NIH). NA Mundell was supported by grants from the NIH, NS083848 (PI, Connie Cepko) and EY023911 (NA Mundell).
Footnote
Provenance: This is a Guest Perspective commissioned by Editor-in-Chief Zhizhuang Joe Zhao (Pathology Graduate Program, University of Oklahoma Health Sciences Center, Oklahoma City, USA).
Conflicts of Interest: The authors have no conflicts of interest to declare.
Comment on: Krishnakumar R, Chen AF, Pantovich MG, et al. FOXD3 Regulates Pluripotent Stem Cell Potential by Simultaneously Initiating and Repressing Enhancer Activity. Cell Stem Cell 2016;18:104-17.
Respuela P, Nikolić M, Tan M, et al. Foxd3 Promotes Exit from Naive Pluripotency through Enhancer Decommissioning and Inhibits Germline Specification. Cell Stem Cell 2016;18:118-33.
References
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, et al. Embryonic stem cell lines derived from human blastocysts. Science 1998;282:1145-7. [Crossref] [PubMed]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006;126:663-76. [Crossref] [PubMed]
- Ying QL, Wray J, Nichols J, et al. The ground state of embryonic stem cell self-renewal. Nature 2008;453:519-23. [Crossref] [PubMed]
- Kojima Y, Kaufman-Francis K, Studdert JB, et al. The transcriptional and functional properties of mouse epiblast stem cells resemble the anterior primitive streak. Cell Stem Cell 2014;14:107-20. [Crossref] [PubMed]
- Hayashi K, Ohta H, Kurimoto K, et al. Reconstitution of the mouse germ cell specification pathway in culture by pluripotent stem cells. Cell 2011;146:519-32. [Crossref] [PubMed]
- Krishnakumar R, Chen AF, Pantovich MG, et al. FOXD3 Regulates Pluripotent Stem Cell Potential by Simultaneously Initiating and Repressing Enhancer Activity. Cell Stem Cell 2016;18:104-17. [Crossref] [PubMed]
- Respuela P, Nikolić M, Tan M, et al. Foxd3 Promotes Exit from Naive Pluripotency through Enhancer Decommissioning and Inhibits Germline Specification. Cell Stem Cell 2016;18:118-33. [Crossref] [PubMed]
- Hayashi K, Surani MA. Self-renewing epiblast stem cells exhibit continual delineation of germ cells with epigenetic reprogramming in vitro. Development 2009;136:3549-56. [Crossref] [PubMed]
- Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell 2009;4:487-92. [Crossref] [PubMed]
- Brons IG, Smithers LE, Trotter MW, et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 2007;448:191-5. [Crossref] [PubMed]
- Tesar PJ, Chenoweth JG, Brook FA, et al. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 2007;448:196-9. [Crossref] [PubMed]
- Hanna LA, Foreman RK, Tarasenko IA, et al. Requirement for Foxd3 in maintaining pluripotent cells of the early mouse embryo. Genes Dev 2002;16:2650-61. [Crossref] [PubMed]
- Liu Y, Labosky PA. Regulation of embryonic stem cell self-renewal and pluripotency by Foxd3. Stem Cells 2008;26:2475-84. [Crossref] [PubMed]
- Mundell NA, Labosky PA. Neural crest stem cell multipotency requires Foxd3 to maintain neural potential and repress mesenchymal fates. Development 2011;138:641-52. [Crossref] [PubMed]
- Plank JL, Suflita MT, Galindo CL, et al. Transcriptional targets of Foxd3 in murine ES cells. Stem Cell Res 2014;12:233-40. [Crossref] [PubMed]
- Suflita MT, Pfaltzgraff ER, Mundell NA, et al. Ground-state transcriptional requirements for skin-derived precursors. Stem Cells Dev 2013;22:1779-88. [Crossref] [PubMed]
- Teng L, Mundell NA, Frist AY, et al. Requirement for Foxd3 in the maintenance of neural crest progenitors. Development 2008;135:1615-24. [Crossref] [PubMed]
- Tompers DM, Foreman RK, Wang Q, et al. Foxd3 is required in the trophoblast progenitor cell lineage of the mouse embryo. Dev Biol 2005;285:126-37. [Crossref] [PubMed]
- Sweet DJ. Foxd3: A Repressor, an Activator, or Both? Cell Stem Cell 2016;18:1-2. [Crossref] [PubMed]
- Mundell NA, Plank JL, LeGrone AW, et al. Enteric nervous system specific deletion of Foxd3 disrupts glial cell differentiation and activates compensatory enteric progenitors. Dev Biol 2012;363:373-87. [Crossref] [PubMed]
Cite this article as: Plank-Bazinet JL, Mundell NA. The paradox of Foxd3: how does it function in pluripotency and differentiation of embryonic stem cells? Stem Cell Investig 2016;3:73.


