Regulatory factors of induced pluripotency: current status
Introduction
Reprogramming is the process by which a differentiated somatic cell reverts to a pluripotent state, from which it can adopt any cellular identity (1,2). During development, cell fate is established and maintained by complex regulatory networks of transcription factors that promote the expression of cell-type specific gene products and repress regulators of other lineages. Despite numerous intrinsic and extrinsic perturbations, cellular identity is remarkably stable once established. This stability is likely the result of a combination of multiple molecular features including cis-acting epigenetic modifications, such as DNA methylation, post-translational modifications of histone tails, nucleosome positioning, incorporation of histone variants into nucleosomes, and trans-acting regulatory factors, such as sequence-specific DNA-binding transcription factors, transcriptional co-activators, non-coding RNAs, and chromatin remodeling complexes (3). Although generally stable in vivo, differentiated cell fate can be dominantly reprogrammed to pluripotent status by various methods (Figure 1A). These methods include: (I) somatic cell nuclear transfer (SCNT); (II) cell fusion; (III) enforced expression of transcription factors [Oct4, Sox2, Klf4, and c-Myc (OSKM)] to generate induced pluripotent stem cells (iPSCs) (4-6). A team of researchers from Japan and Boston reported a cellular reprogramming phenomenon that sublethal stress, such as low pH medium, can induced neonatal somatic cells into pluripotency (STAP, Figure 1B) (7,8). But this method is still controversial. iPSC has less ethical and legal issues than SCNT (5). Moreover, iPSCs offer invaluable sources of patient-specific pluripotent stem cells for disease modeling, drug screening, toxicology tests, and regenerative medicine (9).
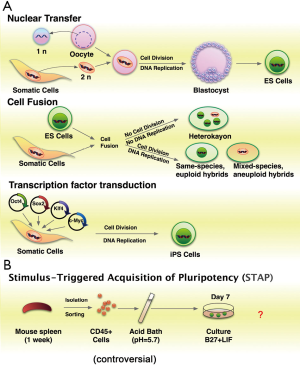
There are, however, several hurdles need to be overcome before iPSCs used in a therapeutic setting (10,11). Currently, iPSC induction is typically slow. The reprogramming of somatic cells from accessible adult tissues is particularly inefficient, because the cells are at a late stage of differentiation (10). Acquisition of induced pluripotency is a slow (usually more than 2 weeks in human) and inefficient (0.1-3%) process. It indicates that cellular reprogramming need to overcome a series of barriers (12). Increased understanding of the molecular and regulatory mechanisms of the reprogramming process is essential to improve the quality of resulting iPSCs for potential therapeutic applications and to address fundamental questions about the control of cell identity (13). In this review, we briefly summarize the current understanding of induced reprogramming and focus on the roles of regulators in this process. We also discuss the future directions of reprogramming research.
Molecular events of OSKM-induced reprogramming
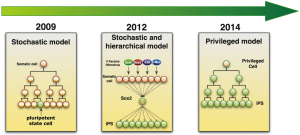
Given the fact that only few starting cells become iPSCs, a number of models have been developed to explain the inefficiency of iPSC generation (Figure 2) (14). In 2009, two contending models were initially proposed, namely the stochastic model and elite model. In the elite model, small numbers of cells are predetermined for reprogramming even before transduction of OSKM. However, in the stochastic model, most differentiated cells have the potential to become iPS cells after OSKM transduction. The cells become pluripotent dependent on “a sufficient push” from proper expression of OSKM and success of overcoming epigenetic block (15). In the same year, Hanna and his colleagues showed that the stochastic model is more favorable to explain the iPS reprogramming process with inducible reprogramming systems (16). Later on the same group modified their model by combining stochastic and hierarchical model. They demonstrated that stochastic events of gene expression were in early stage of reprogramming. It is followed by a late hierarchical phase with Sox2 being the upstream factor in a gene expression hierarchy (17). Therefore, epigenetic priming events early in the reprogramming process might be critical for pluripotency induction (2). However, recently, Tanabe’s group demonstrated that maturation and not initiation is the limiting step during human fibroblast reprogramming (18). Disparities in the reprogramming process between mouse and human cells is likely due to the fact that conventional mouse and human iPSCs represent different states of pluripotency; these cells differ epigenetically as highlighted by their X chromosome inactivation state. Recently, Guo et al. identified a privileged somatic cell state in which acquisition of pluripotency occurred in a non-stochastic manner. They showed that granulocyte monocyte progenitors (GMP, “privileged cells”) are highly efficient in reprogramming. And they think the privileged state is different from the conventional “elite” cells (19).
iPSC reprogramming proceeds in a stepwise manner (2,14). Early works showed that fibroblasts initially reduce somatic state markers and subsequently activate pluripotency genes, suggesting an ordered process. Fully reprogrammed iPSCs activate endogenous pluripotency genes including Oct4, Sox2, and Nanog to acquire a self-sustaining pluripotent state in which exogenous factors are no longer required (12,20,21). Current evidence showed that iPSC reprogramming is a multistep process in which failure to transition through any of the steps would lead to the low overall reprogramming efficiency (2,22). Utilizing specific surface marker combinations, cells poised to become iPSCs can be enriched at different times during reprogramming. For example, Thy1– and SSEA1+ intermediate cells generated iPSCs with significantly higher efficiency compared with Thy1+ and SSEA1– cells (22). O’Malley et al. demonstrated that, in mouse embryonic fibroblasts (MEFs), reprogramming follows an orderly sequence of stage transitions marked by a decrease in CD44 and an increase in ICAM1 expression (23). Similarly, Quintanilla et al. validated that CD44 is a negative cell surface marker for human fibroblast reprogramming (24). These findings improve the understanding of a detailed reprogramming process, and may lead to new reprogramming strategies.
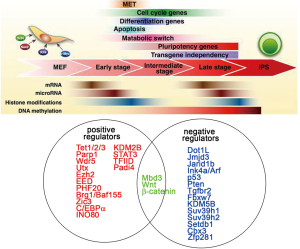
Genome-wide transcriptional profiling has been used to further delineate the sequence of events that drive reprogramming. Initially, cells appear to respond relatively homogeneously to the expression of the reprogramming factors and robustly silence typical mesenchymal genes expressed in fibroblasts, such as Snai1, Snai2, Zeb1, and Zeb2. These events lead to the activation of epithelial markers (such as Cdh1, Epcam, and Ocln) in a process called mesenchymal-to-epithelial transition (MET). MET appears to be critical for the early reprogramming phase and is accompanied by morphological changes, increased proliferation, and the formation of cell clusters (25-27). The key characteristic of the subsequent reprogramming phase is the gradual activation of pluripotency-associated genes. The pluripotency loci Nanog and Sall4 are transcriptionally upregulated at a late intermediate stage, whereas others, such as Utf1 and endogenous Sox2, are induced even later, closely mirroring the acquisition of the full pluripotency expression programming (17,28). Although detailed time course of transcriptional studies describing the stage transitions in reprogramming cells have been performed at the single-cell level, facilitators and inhibitors of reprogramming are not easily identified from these data. Because fundamental changes in gene expression during reprogramming occur at the epigenetic level. In the next section, we focus on the epigenetic regulators of the OSKM-induced reprogramming process (Figure 3).
Epigenetic regulation of iPSC reprogramming
DNA methylation regulators
DNA methylation maintains long-lasting cell memories and therefore, is considered to be a pivotal epigenetic barrier to cellular reprogramming (29,30). High resolution mapping of DNA methylation has revealed an intriguing distribution of methylated cytosine in pluripotent stem cells (31). Intriguingly, DNA hypermethylation at the promoters of tissue-specific genes with low CpG density is accompanied by bivalent chromatin in embryonic stem cells (ESCs) and iPSC. And DNA methylation changes mostly occur at the end of the reprogramming process (22). The inhibition of DNA methylation by chemical compounds or RNA interference that target DNA methyltransferases (Dnmts) can increase the efficiency of iPSC generation (32). These finding indicated that changes in DNA methylation and hydroxymethylation play important roles in genome-wide epigenetic remodeling during reprogramming.
Dnmts in cellular reprogramming
DNA methylation is preserved by the maintenance methyltransferase Dnmt1 and established by the de novo methyltransferases Dnmt3a and Dnmt3b. The loss of Dnmt1 causes the loss of two-thirds of total DNA methylation, thus leading to embryonic lethality (33). Embryos with mutant Dnmt3b appear to be normal in early developmental stages but show multiple developmental defects in the later stages (34). Although the Dnmt family plays an essential role in both developmental and germ cell reprogramming processes, Dnmt3a- and Dnmt3b-mediated de novo methylation is dispensable for iPSC induction (35). Dnmt3b conditional deletion in MEFs leads to a partial loss of DNA methylation (36). Dnmt3a knockdown promotes iPSC formation in human cells, whereas deletion of murine Dnmt3a and Dnmt3b has no consequence on cellular reprogramming (35,37).
Ten-eleven translocations (TETs) in cellular reprogramming
TET proteins have emerged as important regulators of DNA demethylation. TETs catalyze the hydroxylation of 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), which serves as a substrate for thymine DNA glycosylase (TDG)-mediated base excision repair into unmodified cytosine (38). The TET1-binding sites overlap with Polycomb group (PcG) target sites (39). Knockdown of TET1 decreases the expression of PcG target genes and pluripotency-related genes, indicating that gene regulation by TET1 cannot be completely explained by the collaborative functioning with PcG (40). Moreover, TET1 overexpression can replace Oct4 (TSKM induction system) during cellular reprogramming, providing genetic evidence that TET1 contributes to the activation of endogenous pluripotency genes. In the TSKM reprogramming system, Tet1 facilitated endogenous Oct4 demethylation and reactivation through 5hmC conversion (41). Although Tet1 plays important role in cellular reprogramming in vitro, ESCs from TET1 knockout (KO) mice did not show any aberrations in the maintenance of pluripotency. Moreover, TET1 deficient mice are viable and fertile (42). The Mbd3/nucleosome remodeling and deacetylation (NuRD) complexes directly recognize 5hmC and therefore, may control the expression of TET1 target genes. Yildirim et al. showed that Mbd3 knockdown preferentially affected expression of 5hmC-marked genes and Mbd3 preferentially binds to 5hmC relative to 5mC in vitro (43). Depletion of the Mbd3 surprisingly yielded reprogramming efficiencies of up to 100% within days (44), which we will talk about later.
TET2 has been shown to induce hydroxymethylation of key pluripotency genes such as Nanog shortly after OKSM overexpression. Knockdown of TET2 prevents the reprogramming synergy of Nanog with a catalytically deficient mutant of TET1. Genome-wide Chromatin Immunoprecipitation Sequencing (ChIP-seq) analyses further revealed that TET1 and TET2 co-occupy many pluripotency targets in ESCs (45). In agreement with these analyses, Zhu et al. develop a combination of modified reprogramming factors (OySyNyK) which significantly increased Tet1 expression at the early stage and interact with TET1/2 to promote reprogramming (46). Doege et al. identified poly (ADP-ribose) polymerase-1 (Parp1) and TET2 necessary for iPSC generation, which were recruited to the Nanog and Esrrb loci during the early stage of reprogramming. They further showed that Parp1 functioned in the regulation of 5mC modification (47).
Tet1 and Tet2 are highly expressed in mouse ES cells, but Tet3 is more enriched in oocytes and one-cell zygotes (48). Gu et al. showed that Tet3-mediated DNA hydroxylation is involved in epigenetic reprogramming of the zygotic paternal DNA following natural fertilization and may also contribute to somatic cell nuclear reprogramming during animal cloning (49). Recently, TET1/2/3 triple KO MEFs were derived by the same research group. They found that MEFs deleted in all three Tet genes cannot be reprogrammed because of a block in the MET step (50).
Histone-modifying enzymes in reprogramming
Histone marks and chromatin structure are regulated by histone modifying enzymes including “reader”, such as PHD finger proteins, “writers”, such as histone methyltransferases (HMTs) and histone acetyltransferases, and “erasers” such as histone demethylases (HDMs) and histone deacetylases (HDACs) (51). These enzymes function as co-activators or co-repressors of OKSM at different stages of reprogramming and can profoundly influence iPSC derivation (12). Recent technical advances have allowed us to map chromatin modifications throughout the genome by combining ChIP with DNA microarray analysis or high-performance sequencing. These methodologies have revealed that pluripotent stem cells have a unique expression pattern for histone modifiers and distinct distributions of modified histones.
Trimethylation of histone 3 lysine 4 (H3K4), an active marker of transcription, is frequently observed in promoter regions of pluripotent stem cells (52). ESC pluripotency is regulated in part by H3K4 methylation; however, it is still unclear whether H3K4 methylation is involved in iPSC reprogramming. Studies have shown the Trithorax group (TrxG) complexes with the activity of H3K4 methylation to promote iPSC reprogramming, thus providing a functional link between H3K4 methylation and reprogramming. Wdr5, a key component of TrxG, interacts with H3K4me2 to mediate the transition of H3K4me2 to H3K4me3. The expression of Wdr5 is the highest in undifferentiated ESCs and iPSCs and decreases during the differentiation process. Wdr5 has been shown to enhance the efficiency of OSKM-mediated iPSC generation by interacting with Oct4 (53). Kidder et al. found that the H3K4-specific demethylase KDM5B is a barrier to the reprogramming process as evidenced by the accelerated reprogramming of differentiated cells in the absence of KDM5B (54).
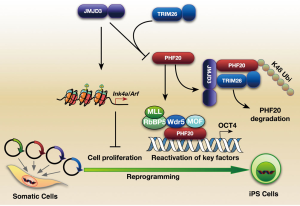
The methylation of histone 3 lysine 27 (H3K27) is mediated by polycomb repressive complex 2 (PRC2), which is composed of PcG proteins such as enhancer of zeste 2 (EZH2), embryonic ectoderm development (EED), and suppressor of zeste 12 homolog (Suz12) (55). Onder et al. used short hairpin RNAs (shRNAs) to target genes in DNA and histone methylation pathways, and identified that EZH2 and EED KD reduced reprogramming efficiency (37). The JmjC domain-containing proteins UTX and JMJD3 demethylate trimethylate H3K27 (56). Our group showed that Jmjd3 blocks reprogramming not only by activating the Ink4a/Arf locus but also by targeting the methyl-lysine effector protein PHF20 for ubiquitination (Figure 4). We also found that PHF20 in collaboration with Wdr5 is required to activate Oct4 transcription (57). Interestingly, Utx promotes somatic and germ cell epigenetic reprogramming. Hanna and his colleagues showed that Utx deficient somatic cells failed to robustly reprogram back to the ground state of pluripotency. Utx directly interacted with OSKM to facilitate iPSC generation in a HDMs activity-independent manner (58).
The histone 3 lysine 9 (H3K9) methyltransferases (HMT) maintain the refractory heterochromatic state of somatic cells, thus acting as major barriers to reprogramming. The H3K9 methyltransferase G9a is essential for embryonic development and has been shown to prevent reprogramming by recruiting Dnmt3a and Dnmt3b to the promoters of Oct4 and HP1β (59). Pei and colleagues show that bone morphogenetic protein (BMP) signaling to H3K9 methylation is a barrier to reprogramming somatic cells into iPSCs. Setdb1 knockdown led to ~100% efficiency in the reprogramming of pre-iPSCs into iPSCs in the presence of vitamin C (60). Consistent with this notion, knockdown of Suv39h1, Suv39h2, Setdb1, or heterochromatin protein-1γ (Cbx3), increase transcription factor accessibility and result in more efficient iPSC generation from somatic cells (37,61).
Activation of the histone 3 lysine 36 (H3K36) demethylases (HDMs), Jhdm1a and Jhdm1b promote intermediate to late stages of iPSC generation by suppressing the Ink4/Arf locus, which is essential for the acquisition of immortality (62,63). An additional early role for Jhdm1b in epithelial gene activation has recently been reported. Liang’s group showed that KDM2B, a histone H3 Lys 36 dimethyl (H3K36me2)-specific demethylase, functioned at the beginning of the reprogramming process and promoted activation of early responsive genes in reprogramming. This capacity depends on its demethylase and DNA-binding activities and is largely independent of its role in antagonizing senescence (64).
Inhibition of the histone 3 lysine 79 (H3K79) HMT DOT1L significantly increased reprogramming efficiency and substituted for KLF4 and c-Myc. Inhibition of DOT1L early in the reprogramming process led to a marked increase of Nanog and LIN28, which play essential functional roles in the enhancement of reprogramming (37). H3K79 methylation plays a critical role in the progression of G1 phase, S phase, mitosis, and meiosis. Depletion of DOT1L results in reduced cell proliferation in mouse ESCs and human cancer cells. By contrast, the cardiac-specific deletion of DOT1L leads to increased proliferation of heart tissues (65). The specific function of DOT1L in cell proliferation of cellular reprogramming requires further elucidation.
Chromatin remodelers also play important role in cellular reprogramming (12). Singhal et al. identified components of the ATP-dependent BAF chromatin-remodeling complex, Brg1 and Baf155, which significantly increases reprogramming efficiency when used together with the four factors (66). Wang et al. showed that INO80 complex, a SWI/SNF family chromatin remodeler facilitates pluripotency gene activation in ESC self-renewal, reprogramming, and blastocyst development. INO80 co-occupied pluripotency gene promoters with the master transcription factors. At the pluripotency genes, INO80 promoted the recruitment of RNA polymerase II complex for gene activation by maintaining open chromatin architecture (67). Ingrid Grummt and colleagues showed that downregulation of the NuRD complex is required for efficient reprogramming. Overexpression of Mbd3, a subunit of NuRD, inhibits induction of iPSCs. Almost at same time, Jacob Hanna group showed that depletion of Mbd3 yielded reprogramming efficiencies of up to 100% within days, suggesting that elimination of a single gene suffices to render reprogramming a deterministic process (44). However, another research team in UK, reported that overexpression of Mbd3/NuRD facilitates reprogramming in a context-dependent manner. Mbd3 not only facilitated the initiation of neural stem cell reprogramming but also was required for efficient iPSC generation from EpiSCs and pre-iPSCs (68). Therefore, deeper investigation is needed to understand the molecular mechanism of Mbd3 in cellular reprogramming.
Non-coding RNAs in reprogramming
To improve the quality of generated iPSCs, researchers have also focused on using non-coding RNAs such as miRNAs, which are associated with regulation of the epigenome. Two groups reported that the transfection of microRNA (miR)-302 and miR-367 clusters successfully reprogramed mouse and human somatic cells to iPSCs without the use of exogenous transcription factors (69,70). Similarly, KO of the miR-302/367 cluster by TALE nucleases (TALENs) completely blocked iPSC generation (71). The molecular mechanism of miR-302 and miR-367 induced pluripotency is via activating endogenous Oct4 and accelerating MET (69,72,73). Moreover, expression of exogenous miR-302 cluster (without miR-367) is efficient in achieving a fully reprogrammed iPS state in partially reprogrammed cells by relieving Mbd2-mediated inhibition of Nanog expression (74).
Many miRNAs have been shown to promote OSKM-induced reprogramming. The miRNAs miR-291-3p, miR-294 and miR-295 increase the efficiency of reprogramming by Oct4, Sox2 and Klf4, but not by these factors plus c-Myc (75). It was also reported that the activation of BMP signaling induced the expression of miR-205 and miR-200 family members and enhanced MET (27). Li et al. systematically studied small RNA-mediated regulation of iPS cell generation by KD miRNAs during cellular reprogramming. They found that miR-17, miR-25, miR-106a and miR-302b clusters were induced during the early stage of reprogramming. And miR-93 and miR-106b enhance iPSC induction and MET step of reprogramming (76).
miRNAs also suppress reprogramming. For example, Yamanaka’s group showed that inhibition of let-7 during reprogramming leads to an increase in the level of the let-7 target LIN-41/TRIM71, which in turn promotes reprogramming and is important for overcoming the let-7 barrier to reprogramming (77). Another important miRNA barrier for reprogramming is the p53-mediated pathway, which induces the expression of miR-34 family members and suppression of the pluripotency factors Nanog and Sox2. Genetic deletion of miR-34a increases the efficiency and kinetics of reprogramming and establishes pluripotency at a late stage (78). Additionally, the suppression of p53 through the overexpression of miR-138 or repression of miR-21 and miR-29a, enhances reprogramming (79,80).
Other signaling pathways and regulators in reprogramming
Tumor suppressor genes have been found to inhibit reprogramming. p53 has been implicated as an enforcer of differentiation by virtue of its ability to limit the cardinal stem cell characteristic of self-renewal in several systems. Dr. Zhao’s team first found that p53 siRNA and undifferentiated embryonic cell transcription factor 1 (UTF1) enhanced the efficiency of iPSC generation up to 100 fold (81). Later, four research teams demonstrated that p53 is a potent reprogramming barrier by promoting cell senescence (63,82-84). Pten is one of most lost tumor suppressors in human cancer. Recently, Liao et al. found that Pten deletion promotes reprogramming of MEFs into iPSCs. They also showed that the Pten inhibitor dipotassium bisperoxo (5-hydroxypyridine-2-carboxyl) oxovanadate could be used to improve the efficiency of germline-competent iPSC derivation from mouse somatic cells (85).
Wnt/β-catenin signaling pathway plays a pivotal role in the maintenance of pluripotency as well as somatic cell reprogramming. As early as 2008, Marson’s team reported that soluble Wnt3a can be used to enhance the efficiency of reprogramming in combination with nuclear factors, Oct4, Sox2 and Klf4 (86). Recently, Zhang et al. found that β-catenin signaling enhances reprogramming efficiency primarily at the initial stage. β-catenin interacts with reprogramming factors Klf4, Oct4 and Sox2, further enhancing expression of pluripotency circuitry genes (87). Another study demonstrated that increase of β-catenin promoted the activity of Oct4 and Nanog, and enhanced pluripotency (88). However, a series of reports indicate that β-catenin may not be required for pluripotent stem cell self-renewal and expansion. It has been reported that OCT4 represses β-catenin signaling during self-renewal and knockdown of OCT4 activates β-catenin signaling in hESCs (89). Ho et al. demonstrated that active Wnt signaling inhibits the early stage of iPSC reprogramming but is required and even stimulated during the late stage (90). These findings suggest that the effect of β-catenin may be context dependent.
It is well known that the TGF superfamily member BMP4 cooperates with leukemia inhibitory factor (LIF) to maintain the pluripotency of mouse ESCs (91). Jeffrey Wrana group highlighted the important role of BMP signaling in promoting the MET stage of reprogramming. BMP could induce miR-205 and miR-200 family of microRNAs to modulate MET (27). Chen group further demonstrated that BMP4 and BMP7 enhanced the expression of epithelial genes (Cdh1, EpCAM, etc.) and inhibit the expression of mesenchymal genes (Zeb1, Twist1, etc.) in OS-infected MEFs (92). In 2009, Maherali and colleagues showed that inhibition of TGF-β signaling enhanced both the efficiency and kinetics of OSKM-reprogrammed MEFs, whereas activation of the TGF-β signaling blocked reprogramming (93). It was further demonstrated that TGF-β inhibitor could replace Sox2 in reprogramming through induction of the transcription factor Nanog (94).
Recent progress in reprogramming research now points to an important role for transcription factors in the establishment and maintenance of pluripotent phenotypes. Yang et al. (95) discovered that STAT3 activation can directly convert epiblast stem cells into naïve iPSCs in 2i medium. They also demonstrated that STAT3 activation plays a vital role in late-stage somatic cell reprogramming (i.e., activation of endogenous Oct4 gene). Thus, STAT3 activity is essential for converting the primed state to naïve pluripotency state in the mouse. Pijnappel et al. showed that knockdown of the transcription factor IID complex affects the pluripotent circuitry in mouse ESCs and inhibits reprogramming of fibroblasts. Transient expression of TFIID subunits greatly enhanced reprogramming (96). Padi4, a member of peptidylarginine deiminases (PADIs), increased during reprogramming in mouse. Padi4 could bind to regulatory elements of key stem cell genes to activate their expression. Padi4 inhibition significantly reduced reprogramming efficiency (97). Similarly, CCAAT/enhancer binding protein-α (C/EBPα) enhanced reprogramming when co-expressed with OSKM. Ectopic expression of C/EBPα is essential in reprogramming of mature B cells (98). Overexpressing C/EBPα with OSKM induces a 100-fold increase in iPS cell reprogramming efficiency. Pluripotency and epithelial-mesenchymal transition (EMT) genes were markedly upregulated during this conversion. Moreover, C/EBPα transiently made the chromatin of pluripotency genes more accessible to DNase I and induced the expression of TET2 after OSKM induction (99). Zinc finger protein of the cerebellum (Zic)3, a member of Gli family of transcription factors is essential for maintaining ESC pluripotency. Declercq et al. showed that MEFs transduced with Zic3 plus OSK enhanced iPSC formation compared with OSK alone. Zic3 also enhanced the expression of genes known to enhance iPSCs derivation including Nanog and Tbx3 (100). Fidalgo et al. identified Zfp281as a roadblock to efficient somatic cell reprogramming. Zfp281 recruited the NuRD repressor complex onto the Nanog locus and mediated Nanog transcription in repression (101).
Many other signaling pathways are also reported to regulate reprogramming. For example, protein ubiquitination system (UPS) mediates the rapid and highly specific degradation of intracellular proteins and thereby, contributes to the dynamic regulation of protein abundance. Using UPS-targeted RNA interference screens, Buckley et al. identified a significant number of ubiquitin enzymes essential in pluripotency regulation, including Psmd14 and Fbxw7. Psmd14 expression is regulated during ESC differentiation and its silencing affects ESC pluripotency and abrogates cellular reprogramming. On the other hand, the depletion of E3 ligase Fbxw7 leads to the inhibition of differentiation and enhancement of iPSC generation (102). Interestingly, biochemical and biophysical factors can also help reprogram somatic cells into pluripotent stem cells. Downing et al. showed that parallel microgrooves on the surface of cell-adhesive substrates can replace the effects of small molecule epigenetic modifiers and significantly improve reprogramming efficiency (103).
Small chemicals to promote OSKM-induced reprogramming
Stem cell fate is regulated by both intrinsic/extrinsic regulators and the extracellular niche. Because these regulators have limitations in their efficiency and selectivity for controlling stem cell fate, a new strategy is to use small molecules. Surprisingly, a research team lead by Hou showed that pluripotent stem cells can be generated from mouse somatic cells at a frequency up to 0.2% using a combination of seven small molecule compounds (104). Before that, Esteban and colleagues show that vitamin C enhances iPSC generation from both mouse and human somatic cells (105). They further showed that vitamin C induced H3K36me2/3 demethylation during reprogramming and identified KDM2A/2B, two known vitamin-C-dependent H3K36 demethylases, as potent regulators of reprogramming (62). PD0325901 (mitogen-activated protein kinase kinase inhibitor) and CHIR99021 (glycogen synthase kinase-3 inhibitor) (2i) plus LIF have been shown to induced stable up-regulation of Oct4 and Nanog, reactivation of the X chromosome, transgene silencing, and competence for somatic and germline chimaerism (106). The Rho-associated kinase inhibitors Y-27632 and thiazovivin enhance the survival of human ESCs, whereas a combination of PD0325901, CHIR99021, and Y-27632 supplemented with basic fibroblast growth factor supports the maintenance of human ESCs (107). Recently, Hanna and colleagues have established defined conditions that facilitate the derivation of human naïve ground state pluripotent stem cells with a chemical cocktail (108). Compared to genetic manipulations, small molecule approaches have a number of advantages: (I) the biological effects of small molecules are rapid, reversible, and dose-dependent; (II) small molecules will not cause instability of genome; and (III) a variety of chemical libraries provide data for the optimization of small molecule effects. Small chemicals could be useful for the iPS technology in clinic.
Future directions
The medical applications of human iPSCs in disease modeling and stem cell therapy have been progressing rapidly. Elucidation of the details and mechanisms of the reprogramming process during iPSC generation has resolved many problems regarding the clinical use of iPSCs. The first human clinical trial using autologous iPSCs has been approved by the Japan Ministry Health and will be conducted in 2014 in Kobe. Although the clinical application of iPSC technology has a bright future, challenges remain including concerns regarding the safety of OSKM-induced reprogramming. Numerous alternative methods for inducing pluripotency without the use of viral vectors have been reported, but their efficiency remains problematic (109). In conclusion, we need to identify more deterministic regulators, in particular the small chemicals, which lead to global changes in the epigenetic regulation of somatic cells from a differentiated state to a pluripotent state. Further research is needed to efficiently generate high-quality and safe iPSCs for clinical use.
Acknowledgements
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Takahashi K. Cellular reprogramming. Cold Spring Harb Perspect Biol 2014.6. [PubMed]
- Papp B, Plath K. Epigenetics of reprogramming to induced pluripotency. Cell 2013;152:1324-43. [PubMed]
- Orkin SH, Hochedlinger K. Chromatin connections to pluripotency and cellular reprogramming. Cell 2011;145:835-50. [PubMed]
- Yamanaka S, Blau HM. Nuclear reprogramming to a pluripotent state by three approaches. Nature 2010;465:704-12. [PubMed]
- Stadtfeld M, Hochedlinger K. Induced pluripotency: history, mechanisms, and applications. Genes Dev 2010;24:2239-63. [PubMed]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006;126:663-76. [PubMed]
- Obokata H, Wakayama T, Sasai Y, et al. Stimulus-triggered fate conversion of somatic cells into pluripotency. Nature 2014;505:641-7. [PubMed]
- Obokata H, Sasai Y, Niwa H, et al. Bidirectional developmental potential in reprogrammed cells with acquired pluripotency. Nature 2014;505:676-80. [PubMed]
- Cherry AB, Daley GQ. Reprogrammed cells for disease modeling and regenerative medicine. Annu Rev Med 2013;64:277-90. [PubMed]
- Skene PJ, Henikoff S. Chromatin roadblocks to reprogramming 50 years on. BMC Biol 2012;10:83. [PubMed]
- Vierbuchen T, Wernig M. Molecular roadblocks for cellular reprogramming. Mol Cell 2012;47:827-38. [PubMed]
- Apostolou E, Hochedlinger K. Chromatin dynamics during cellular reprogramming. Nature 2013;502:462-71. [PubMed]
- Buganim Y, Faddah DA, Jaenisch R. Mechanisms and models of somatic cell reprogramming. Nat Rev Genet 2013;14:427-39. [PubMed]
- Theunissen TW, Jaenisch R. Molecular Control of Induced Pluripotency. Cell Stem Cell 2014;14:720-734. [PubMed]
- Yamanaka S. Elite and stochastic models for induced pluripotent stem cell generation. Nature 2009;460:49-52. [PubMed]
- Hanna J, Saha K, Pando B, et al. Direct cell reprogramming is a stochastic process amenable to acceleration. Nature 2009;462:595-601. [PubMed]
- Buganim Y, Faddah DA, Cheng AW, et al. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell 2012;150:1209-22. [PubMed]
- Tanabe K, Nakamura M, Narita M, et al. Maturation, not initiation, is the major roadblock during reprogramming toward pluripotency from human fibroblasts. Proc Natl Acad Sci U S A 2013;110:12172-9. [PubMed]
- Guo S, Zi X, Schulz VP, et al. Nonstochastic reprogramming from a privileged somatic cell state. Cell 2014;156:649-62. [PubMed]
- Brambrink T, Foreman R, Welstead GG, et al. Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell 2008;2:151-9. [PubMed]
- Stadtfeld M, Maherali N, Breault DT, et al. Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell 2008;2:230-40. [PubMed]
- Polo JM, Anderssen E, Walsh RM, et al. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell 2012;151:1617-32. [PubMed]
- O’Malley J, Skylaki S, Iwabuchi KA, et al. High-resolution analysis with novel cell-surface markers identifies routes to iPS cells. Nature 2013;499:88-91. [PubMed]
- Quintanilla RH Jr, Asprer JS, Vaz C, et al. CD44 is a negative cell surface marker for pluripotent stem cell identification during human fibroblast reprogramming. PLoS One 2014;9:e85419. [PubMed]
- Papp B, Plath K. Reprogramming to pluripotency: stepwise resetting of the epigenetic landscape. Cell Res 2011;21:486-501. [PubMed]
- Li R, Liang J, Ni S, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell 2010;7:51-63. [PubMed]
- Samavarchi-Tehrani P, Golipour A, David L, et al. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell 2010;7:64-77. [PubMed]
- Plath K, Lowry WE. Progress in understanding reprogramming to the induced pluripotent state. Nat Rev Genet 2011;12:253-65. [PubMed]
- De Carvalho DD, You JS, Jones PA. DNA methylation and cellular reprogramming. Trends Cell Biol 2010;20:609-17. [PubMed]
- Huang K, Fan G. DNA methylation in cell differentiation and reprogramming: an emerging systematic view. Regen Med 2010;5:531-44. [PubMed]
- Meissner A, Mikkelsen TS, Gu H, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature 2008;454:766-70. [PubMed]
- Okita K, Yamanaka S. Induced pluripotent stem cells: opportunities and challenges. Philos Trans R Soc Lond B Biol Sci 2011;366:2198-207. [PubMed]
- Biniszkiewicz D, Gribnau J, Ramsahoye B, et al. Dnmt1 overexpression causes genomic hypermethylation, loss of imprinting, and embryonic lethality. Mol Cell Biol 2002;22:2124-35. [PubMed]
- Ueda Y, Okano M, Williams C, et al. Roles for Dnmt3b in mammalian development: a mouse model for the ICF syndrome. Development 2006;133:1183-92. [PubMed]
- Pawlak M, Jaenisch R. De novo DNA methylation by Dnmt3a and Dnmt3b is dispensable for nuclear reprogramming of somatic cells to a pluripotent state. Genes Dev 2011;25:1035-40. [PubMed]
- Dodge JE, Okano M, Dick F, et al. Inactivation of Dnmt3b in mouse embryonic fibroblasts results in DNA hypomethylation, chromosomal instability, and spontaneous immortalization. J Biol Chem 2005;280:17986-91. [PubMed]
- Onder TT, Kara N, Cherry A, et al. Chromatin-modifying enzymes as modulators of reprogramming. Nature 2012;483:598-602. [PubMed]
- Ito S, D’Alessio AC, Taranova OV, et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 2010;466:1129-33. [PubMed]
- Williams K, Christensen J, Pedersen MT, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature 2011;473:343-8. [PubMed]
- Watanabe A, Yamada Y, Yamanaka S. Epigenetic regulation in pluripotent stem cells: a key to breaking the epigenetic barrier. Philos Trans R Soc Lond B Biol Sci 2013;368:20120292. [PubMed]
- Gao Y, Chen J, Li K, et al. Replacement of Oct4 by Tet1 during iPSC induction reveals an important role of DNA methylation and hydroxymethylation in reprogramming. Cell Stem Cell 2013;12:453-69. [PubMed]
- Dawlaty MM, Ganz K, Powell BE, et al. Tet1 is dispensable for maintaining pluripotency and its loss is compatible with embryonic and postnatal development. Cell Stem Cell 2011;9:166-75. [PubMed]
- Yildirim O, Li R, Hung JH, et al. Mbd3/NURD complex regulates expression of 5-hydroxymethylcytosine marked genes in embryonic stem cells. Cell 2011;147:1498-510. [PubMed]
- Rais Y, Zviran A, Geula S, et al. Deterministic direct reprogramming of somatic cells to pluripotency. Nature 2013;502:65-70. [PubMed]
- Costa Y, Ding J, Theunissen TW, et al. NANOG-dependent function of TET1 and TET2 in establishment of pluripotency. Nature 2013;495:370-4. [PubMed]
- Zhu G, Li Y, Zhu F, et al. Coordination of Engineered Factors with TET1/2 Promotes Early-Stage Epigenetic Modification during Somatic Cell Reprogramming. Stem Cell Reports 2014;2:253-61. [PubMed]
- Doege CA, Inoue K, Yamashita T, et al. Early-stage epigenetic modification during somatic cell reprogramming by Parp1 and Tet2. Nature 2012;488:652-5. [PubMed]
- Jin SG, Jiang Y, Qiu R, et al. 5-Hydroxymethylcytosine is strongly depleted in human cancers but its levels do not correlate with IDH1 mutations. Cancer Res 2011;71:7360-5. [PubMed]
- Gu TP, Guo F, Yang H, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature 2011;477:606-10. [PubMed]
- Hu X, Zhang L, Mao SQ, et al. Tet and TDG mediate DNA demethylation essential for mesenchymal-to-epithelial transition in somatic cell reprogramming. Cell Stem Cell 2014;14:512-22. [PubMed]
- Sanchez R, Zhou MM. The PHD finger: a versatile epigenome reader. Trends Biochem Sci 2011;36:364-72. [PubMed]
- Mikkelsen TS, Ku M, Jaffe DB, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 2007;448:553-60. [PubMed]
- Ang YS, Tsai SY, Lee DF, et al. Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell 2011;145:183-97. [PubMed]
- Kidder BL, Hu G, Yu ZX, et al. Extended self-renewal and accelerated reprogramming in the absence of Kdm5b. Mol Cell Biol 2013;33:4793-810. [PubMed]
- Boyer LA, Plath K, Zeitlinger J, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 2006;441:349-53. [PubMed]
- Agger K, Cloos PA, Christensen J, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 2007;449:731-4. [PubMed]
- Zhao W, Li Q, Ayers S, et al. Jmjd3 inhibits reprogramming by upregulating expression of INK4a/Arf and targeting PHF20 for ubiquitination. Cell 2013;152:1037-50. [PubMed]
- Mansour AA, Gafni O, Weinberger L, et al. The H3K27 demethylase Utx regulates somatic and germ cell epigenetic reprogramming. Nature 2012;488:409-13. [PubMed]
- Epsztejn-Litman S, Feldman N, Abu-Remaileh M, et al. De novo DNA methylation promoted by G9a prevents reprogramming of embryonically silenced genes. Nat Struct Mol Biol 2008;15:1176-83. [PubMed]
- Chen J, Liu H, Liu J, et al. H3K9 methylation is a barrier during somatic cell reprogramming into iPSCs. Nat Genet 2013;45:34-42. [PubMed]
- Soufi A, Donahue G, Zaret KS. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell 2012;151:994-1004. [PubMed]
- Wang T, Chen K, Zeng X, et al. The histone demethylases Jhdm1a/1b enhance somatic cell reprogramming in a vitamin-C-dependent manner. Cell Stem Cell 2011;9:575-87. [PubMed]
- Li H, Collado M, Villasante A, et al. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature 2009;460:1136-9. [PubMed]
- Liang G, He J, Zhang Y. Kdm2b promotes induced pluripotent stem cell generation by facilitating gene activation early in reprogramming. Nat Cell Biol 2012;14:457-66. [PubMed]
- Barry ER, Krueger W, Jakuba CM, et al. ES cell cycle progression and differentiation require the action of the histone methyltransferase Dot1L. Stem Cells 2009;27:1538-47. [PubMed]
- Singhal N, Graumann J, Wu G, et al. Chromatin-Remodeling Components of the BAF Complex Facilitate Reprogramming. Cell 2010;141:943-55. [PubMed]
- Wang L, Du Y, Ward JM, et al. INO80 facilitates pluripotency gene activation in embryonic stem cell self-renewal, reprogramming, and blastocyst development. Cell Stem Cell 2014;14:575-91. [PubMed]
- Dos Santos RL, Tosti L, Radzisheuskaya A, et al. MBD3/NuRD Facilitates Induction of Pluripotency in a Context-Dependent Manner. Cell Stem Cell 2014;15:102-10. [PubMed]
- Anokye-Danso F, Trivedi CM, Juhr D, et al. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell 2011;8:376-88. [PubMed]
- Miyoshi N, Ishii H, Nagano H, et al. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell 2011;8:633-8. [PubMed]
- Zhang Z, Xiang D, Heriyanto F, et al. Dissecting the Roles of miR-302/367 Cluster in Cellular Reprogramming Using TALE-based Repressor and TALEN. Stem Cell Reports 2013;1:218-25. [PubMed]
- Liao B, Bao X, Liu L, et al. MicroRNA cluster 302-367 enhances somatic cell reprogramming by accelerating a mesenchymal-to-epithelial transition. J Biol Chem 2011;286:17359-64. [PubMed]
- Subramanyam D, Lamouille S, Judson RL, et al. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat Biotechnol 2011;29:443-8. [PubMed]
- Lee MR, Prasain N, Chae HD, et al. Epigenetic regulation of NANOG by miR-302 cluster-MBD2 completes induced pluripotent stem cell reprogramming. Stem Cells 2013;31:666-81. [PubMed]
- Judson RL, Babiarz JE, Venere M, et al. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol 2009;27:459-61. [PubMed]
- Li Z, Yang CS, Nakashima K, et al. Small RNA-mediated regulation of iPS cell generation. EMBO J 2011;30:823-34. [PubMed]
- Worringer KA, Rand TA, Hayashi Y, et al. The let-7/LIN-41 pathway regulates reprogramming to human induced pluripotent stem cells by controlling expression of prodifferentiation genes. Cell Stem Cell 2014;14:40-52. [PubMed]
- Choi YJ, Lin CP, Ho JJ, et al. miR-34 miRNAs provide a barrier for somatic cell reprogramming. Nat Cell Biol 2011;13:1353-60. [PubMed]
- Ye D, Wang G, Liu Y, et al. MiR-138 promotes induced pluripotent stem cell generation through the regulation of the p53 signaling. Stem Cells 2012;30:1645-54. [PubMed]
- Yang CS, Li Z, Rana TM. microRNAs modulate iPS cell generation. RNA 2011;17:1451-60. [PubMed]
- Zhao Y, Yin X, Qin H, et al. Two supporting factors greatly improve the efficiency of human iPSC generation. Cell Stem Cell 2008;3:475-9. [PubMed]
- Kawamura T, Suzuki J, Wang YV, et al. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature 2009;460:1140-4. [PubMed]
- Utikal J, Polo JM, Stadtfeld M, et al. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature 2009;460:1145-8. [PubMed]
- Marión RM, Strati K, Li H, et al. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature 2009;460:1149-53. [PubMed]
- Liao J, Marumoto T, Yamaguchi S, et al. Inhibition of PTEN tumor suppressor promotes the generation of induced pluripotent stem cells. Mol Ther 2013;21:1242-50. [PubMed]
- Marson A, Foreman R, Chevalier B, et al. Wnt signaling promotes reprogramming of somatic cells to pluripotency. Cell Stem Cell 2008;3:132-5. [PubMed]
- Zhang P, Chang WH, Fong B, et al. Regulation of induced pluripotent stem (iPS) cell induction by Wnt/β-catenin signaling. J Biol Chem 2014;289:9221-32. [PubMed]
- Faunes F, Hayward P, Descalzo SM, et al. A membrane-associated β-catenin/Oct4 complex correlates with ground-state pluripotency in mouse embryonic stem cells. Development 2013;140:1171-83. [PubMed]
- Davidson KC, Adams AM, Goodson JM, et al. Wnt/β-catenin signaling promotes differentiation, not self-renewal, of human embryonic stem cells and is repressed by Oct4. Proc Natl Acad Sci U S A 2012;109:4485-90. [PubMed]
- Ho R, Papp B, Hoffman JA, et al. Stage-specific regulation of reprogramming to induced pluripotent stem cells by Wnt signaling and T cell factor proteins. Cell Rep 2013;3:2113-26. [PubMed]
- Qi X, Li TG, Hao J, et al. BMP4 supports self-renewal of embryonic stem cells by inhibiting mitogen-activated protein kinase pathways. Proc Natl Acad Sci U S A 2004;101:6027-32. [PubMed]
- Chen J, Liu J, Yang J, et al. BMPs functionally replace Klf4 and support efficient reprogramming of mouse fibroblasts by Oct4 alone. Cell Res 2011;21:205-12. [PubMed]
- Maherali N, Hochedlinger K. Tgfbeta signal inhibition cooperates in the induction of iPSCs and replaces Sox2 and cMyc. Curr Biol 2009;19:1718-23. [PubMed]
- Ichida JK, Blanchard J, Lam K, et al. A small-molecule inhibitor of tgf-Beta signaling replaces sox2 in reprogramming by inducing nanog. Cell Stem Cell 2009;5:491-503. [PubMed]
- Yang J, van Oosten AL, Theunissen TW, et al. Stat3 activation is limiting for reprogramming to ground state pluripotency. Cell Stem Cell 2010;7:319-28. [PubMed]
- Pijnappel WW, Esch D, Baltissen MP, et al. A central role for TFIID in the pluripotent transcription circuitry. Nature 2013;495:516-9. [PubMed]
- Christophorou MA, Castelo-Branco G, Halley-Stott RP, et al. Citrullination regulates pluripotency and histone H1 binding to chromatin. Nature 2014;507:104-8. [PubMed]
- Hanna J, Markoulaki S, Schorderet P, et al. Direct reprogramming of terminally differentiated mature B lymphocytes to pluripotency. Cell 2008;133:250-64. [PubMed]
- Di Stefano B, Sardina JL, van Oevelen C, et al. C/EBPα poises B cells for rapid reprogramming into induced pluripotent stem cells. Nature 2014;506:235-9. [PubMed]
- Declercq J, Sheshadri P, Verfaillie CM, et al. Zic3 enhances the generation of mouse induced pluripotent stem cells. Stem Cells Dev 2013;22:2017-25. [PubMed]
- Fidalgo M, Faiola F, Pereira CF, et al. Zfp281 mediates Nanog autorepression through recruitment of the NuRD complex and inhibits somatic cell reprogramming. Proc Natl Acad Sci U S A 2012;109:16202-7. [PubMed]
- Buckley SM, Aranda-Orgilles B, Strikoudis A, et al. Regulation of pluripotency and cellular reprogramming by the ubiquitin-proteasome system. Cell Stem Cell 2012;11:783-98. [PubMed]
- Downing TL, Soto J, Morez C, et al. Biophysical regulation of epigenetic state and cell reprogramming. Nat Mater 2013;12:1154-62. [PubMed]
- Hou P, Li Y, Zhang X, et al. Pluripotent stem cells induced from mouse somatic cells by small-molecule compounds. Science 2013;341:651-4. [PubMed]
- Esteban MA, Wang T, Qin B, et al. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell 2010;6:71-9. [PubMed]
- Silva J, Barrandon O, Nichols J, et al. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol 2008;6:e253. [PubMed]
- Valamehr B, Abujarour R, Robinson M, et al. A novel platform to enable the high-throughput derivation and characterization of feeder-free human iPSCs. Sci Rep 2012;2:213. [PubMed]
- Gafni O, Weinberger L, Mansour AA, et al. Derivation of novel human ground state naive pluripotent stem cells. Nature 2013;504:282-6. [PubMed]
- Zhou YY, Zeng F. Integration-free methods for generating induced pluripotent stem cells. Genomics Proteomics Bioinformatics 2013;11:284-7. [PubMed]
Cite this article as: Zhao W, Ning B, Qian C. Regulatory factors of induced pluripotency: current status. Stem Cell Investig 2014;1:15.


