
Seeking fate—CRISPRa screens reveal new neural lineage and reprogramming factors
It is still one of the most intriguing questions in biology, how the multitudes of cell types a single organism possesses, are adequately born during development. It is clear that there are transcription factors with sufficient activity to drive cells towards specific cell identities, however to date the known genes are limited to a handful of linages (e.g., pluripotency, neurogenesis, β-cell development, etc.) and even in these cases we are far from a comprehensive understanding of contributing factors and molecular mechanisms. The reason for this lies at least in part in experimental limitations. Explorative approaches are often dependent on elaborate cDNA expression constructs and thus strongly limited by the number of genes testable. Most studies consequently focused on canonical transcription factors with cell type specific expression patterns. A recent publication by the group of Liu et al. shows however that new less biased approaches can be used for the systematic identification of neurogenic factors (1).
The authors employed a synthetic complex based on the bacterial phage defense system clustered regularly interspaced short palindromic repeats (CRISPR) (2). CRISPR depends on two components, a protein component, the nuclease Cas9 that targets specific genomic sequences encoded in an RNA component, the noncoding guide or gRNA. Since gRNAs are short (~96 bp), they can be easily combined into libraries of viruses expressing defined sets of CRISPR targeting sites (3). Importantly, by employing a nuclease deficient version of Cas9 (dCas9) (4), a large number of effectors can be targeted to chromatin (5). One example is the combination of trans-activating domains with dCas9 (CRISPRa) either through direct fusion of protein components or by the use of protein tags. Among the most versatile dCas9 tags is the so-called SunTag, a short protein sequence allowing targeted gene activation when combined with synthetic antibodies fused to trans-activation domains (6). Similar CRISPRa systems have been used already to activate known neuronal promoting genes (NGN2, NEUROD1), resulting in a higher number of neurons born during embryonic stem cell (ESC) differentiation (7,8).
In their screen Liu and colleagues employ clonal mouse ESC lines expressing the CRISPRa-SunTag system when doxycycline is added to the medium (CRISPR activating mouse ESCs). In order to identify and isolate differentiated neurons, they additionally inserted a human CD8 (hCD8) antigen sequence into the neuronal gene Tubb3. This strategy enables magnetic-activated cell sorting (MACS) of cells expressing the neuronal marker Tuj1. To conduct a comprehensive dCas9 screen they employed a viral library containing a pool of 55,561 gRNAs targeting 2,428 genes that represent the entirety of computationally predicted transcription and DNA binding factors. After 12 days of differentiation, next generation sequencing (NGS) of sorted cells allowed for the identification of 74 hits using a newly developed algorithm. Several of those candidate neuronal promoting genes were already implicated in this process (e.g., Brn2), while notably quite some others were new and often did not even show differential expression during neural development.
The authors went on to validate the 20 top hits using two independent approaches; the transduction of individual gRNAs, confirming 19 out of 20 candidates (including Ngn1, Ezh2, Jun and Suz12), and cDNA over-expression of the respective targets, in this way 14 of 20 were confirmed (excluding Sin3b, Rb1 and others). Although this difference could be due to a number of reasons it might be worth mentioning that a similar phenomenon has been described before (9); induction of the endogenous Sox2 gene has been reported to be more potent in inducing pluripotency than forced expression of cDNA. While it is far too early to conclude that this indicates a shift in paradigms, it could indicate that constitutive overexpression of transgenic constructs might sometimes be detrimental.
For the investigation of subtype or regional specificity of the reprogrammed neurons, Liu et al. analyzed the transcriptome of neurons generated via four different top candidates (Jun, Ngn1, Ezh2, Suz12) and the combination of Ngn1 and Ezh2. Neurons generated by these different approaches shared a common set of upregulated genes that were enriched for factors associated to neural development. Indeed resulting neurons share some transcriptional proximity to in vivo neurons. Even though unique sets of upregulated genes can be seen, hinting towards different transcriptional signatures for the respective neuronal promoting factors, they confirmed the upregulation of glutamatergic programs in all approaches, indicating a tendency towards excitatory neurons. Furthermore, neurons generated by induction of two candidate neuronal promoting factors, Ngn1 or Ezh2, were investigated for their electrophysiological properties in detail. Differentiated neurons were found to generate action potentials upon current induction and exhibit the presence of outward potassium and inward sodium channels. In addition, spontaneous excitatory postsynaptic currents (EPSCs) were observed. Taken together, this data indicates that the new discovered neuronal promoting factors can direct ESC differentiation towards neurogenesis, and those newly generated neurons form functional synapses.
There are clear advantages of using CRISPRa over cDNA expression vectors; one is that it is simpler to activate combinations of genes, another that it is easier to activate those to different degree. Liu and colleagues made use of these advantages and combined each of the candidate neuronal promoting factors with each other. In this way they were able to investigate which factors synergize and which ones are counteractive. A screen was performed as before, and the relative abundance of each combination was calculated in the neuronal and non-neuronal population respectively. These experiments revealed a synergistic cluster of 6 core genes (Ngn1, Zeb1, Tcf15, Foxo1, Ezh2 and Brn2), while many others behaved indifferent to partners or even were counteractive. Moreover, using different gRNAs with variable potency for each gene, they were able to correlate synergistic interactions to induction levels. While for some genes (e.g., Ngn1, Ezh2) interactions were weakened by strong activation, indicating that their high expression leads to saturated neuronal differentiation, others (e.g., Brn2) behaved the other way around, hinting that they might be limiting during neuronal differentiation.
Finally, the authors investigated how strong these newly identified factors are in promoting neuronal cell identities not only from ESCs (i.e., programming), but also from terminally differentiated cells (i.e., reprogramming). For this they tested combination of those factors in direct reprogramming approaches using mouse embryonic fibroblasts (MEFs). In contrast to directed ESC differentiation (used during CRISPRa screening), direct reprogramming depends on cell identity changes beyond the natural potency of the starting cell. Several reprogramming factors potent enough to establish new cell identities and overwrite existing one are known, exemplified first for myocyte reprogramming (MyoD) (10) and pluripotency induction (Oct4, Sox2, Myc, Klf4) (11), but later also for direct neuronal reprogramming using the so-called BAM factors (Brn2, Ascl1, Myt1l) (12). Liu and colleagues transduced cDNA expression constructs of their newly discovered neuronal promoting factors into MEFs. With regard to single factors only Ngn1 showed reprogramming capacity, and only with small efficiency (1% of MEFs induce the neuronal marker Map2). In contrast to single factors however, the investigation of 15 combinations of candidate factors revealed three combinations that worked decidedly better than the canonical reprogramming factor cocktail BAM. Strikingly, these combinations always included Ngn1 and one of three other factors (Brn2, Ezh2, Foxo1). Thus, these factors exhibit a non-linear, highly efficient synergy and increased the reprogramming efficiency from 1% to over 80% in the MEF background. Ezh2 and Mecom, two factors that did not lead to any reprogrammed neurons on their own, also produced small amounts of Map2 positive cells when used in combination. Similarly to the neurons generated by directed ESC differentiation, reprogrammed neurons (using combinations of Ngn1 and Ezh2 or Brn2 and Ezh2) resemble those reprogrammed with established factors in their transcriptomic and electrophysiological properties.
These data collectively show how powerful CRISPRa screens can be to reveal new cell identity factors (Figure 1). It might be however also informative to turn one’s attention to those genes not among the hits. Although among the BAM factors Brn2 has been retrieved as one of the most significant hits (and Ascl1 and Ngn2 have been excluded from the gRNA library), those are not the only neuronal promoting factors known. Quite contrary a significant number of transcription factors have been reported to direct and/or induce a neuronal identity [summarized exhaustively by Masserdotti and colleagues (13)]. Among those factors not found in the CRISPRa screen are repressors of non-neuronal identities [e.g., Myt1l (14)], neuronal progenitor and stem cell factors [e.g., Sox2, Pax6 (15)] as well as strong direct reprogramming factors [e.g., Nurr1, Dlx2, NeuroD4, NeuroD1 (16)]. Although there could be many reasons for their absence, one might be that those factors were not sufficiently induced during CRISPRa screening. Indeed, it has been recently shown for another neural master transcription factor, Sox1, that CRISPRa is not sufficient for significant induction of this gene. Chromatin barriers and especially DNA methylation interfere with the response to targeted transactivation (17). Thus, investigating the drop-out rate in CRISPRa screens would be highly informative.
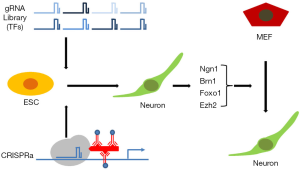
Direct reprogramming is a promising approach for supplying future needs for cell replacement therapies diseases, until now however it lacks efficiency and specificity, hindering its application (18). Thus, it is clear that more has to be learned about the cellular programs that regulate cell identity. Liu and colleagues show that CRISPRa screens are a powerful tool to discover new key players of cell identity. They show that this approach allows the discovery of lineage promoting gene activities without a priori assumptions, even for those lacking cell-specific expression. Moreover, they report one of the first large-scale CRISPRa screen for a complicated and physiologically relevant phenotypic read-out (1). These results open up a new strategy studying cell identity in general, as it is in theory adaptable to any cellular system.
Acknowledgments
The authors like to thank Dr. Giacomo Masserdotti for advice and help with the manuscript.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Liu Y, Yu C, Daley TP, et al. CRISPR activation screens systematically identify factors that drive neuronal fate and reprogramming. Cell Stem Cell 2018;23:758-71.e8. [Crossref] [PubMed]
- Deltcheva E, Chylinski K, Sharma CM, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 2011;471:602-7. [Crossref] [PubMed]
- Shalem O, Sanjana NE, Zhang F. High-throughput functional genomics using CRISPR-Cas9. Nat Rev Genet 2015;16:299-311. [Crossref] [PubMed]
- Gilbert LA, Larson MH, Morsut L, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 2013;154:442-51. [Crossref] [PubMed]
- Stricker SH, Köferle A, Beck S. From profiles to function in epigenomics. Nat Rev Genet 2017;18:51-66. [Crossref] [PubMed]
- Tanenbaum ME, Gilbert LA, Qi LS, et al. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell 2014;159:635-46. [Crossref] [PubMed]
- Chavez A, Scheiman J, Vora S, et al. Highly efficient Cas9-mediated transcriptional programming. Nat Methods 2015;12:326-8. [Crossref] [PubMed]
- Shao J, Wang M, Yu G, et al. Synthetic far-red light-mediated CRISPR-dCas9 device for inducing functional neuronal differentiation. Proc Natl Acad Sci U S A 2018;115:E6722-E6730. [Crossref] [PubMed]
- Liu P, Chen M, Liu Y, et al. CRISPR-based chromatin remodeling of the endogenous Oct4 or Sox2 locus enables reprogramming to pluripotency. Cell Stem Cell 2018;22:252-61.e4. [Crossref] [PubMed]
- Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 1987;51:987-1000. [Crossref] [PubMed]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006;126:663-76. [Crossref] [PubMed]
- Vierbuchen T, Ostermeier A, Pang ZP, et al. Direct conversion of fibroblasts to functional neurons by defined factors. Nature 2010;463:1035-41. [Crossref] [PubMed]
- Masserdotti G, Gascón S, Götz M. Direct neuronal reprogramming: learning from and for development. Development 2016;143:2494-510. [Crossref] [PubMed]
- Mall M, Kareta MS, Chanda S, et al. Myt1l safeguards neuronal identity by actively repressing many non-neuronal fates. Nature 2017;544:245-9. [Crossref] [PubMed]
- Heins N, Malatesta P, Cecconi F, et al. Glial cells generate neurons: the role of the transcription factor Pax6. Nat Neurosci 2002;5:308-15. [Crossref] [PubMed]
- Pang ZP, Yang N, Vierbuchen T, et al. Induction of human neuronal cells by defined transcription factors. Nature 2011;476:220-3. [Crossref] [PubMed]
- Baumann V, Wiesbeck M, Breunig CT, et al. Targeted removal of epigenetic barriers during transcriptional reprogramming. Nat Commun 2019;10:2119. [Crossref] [PubMed]
- Ninkovic J, Götz M. Understanding direct neuronal reprogramming-from pioneer factors to 3D chromatin. Curr Opin Genet Dev 2018;52:65-9. [Crossref] [PubMed]
Cite this article as: Baumann V, Stricker SH. Seeking fate—CRISPRa screens reveal new neural lineage and reprogramming factors. Stem Cell Investig 2019;6:30.


