
Lineage switch from T-cell lymphoblastic leukemia/lymphoma to acute myeloid leukemia and back to T-cell lymphoblastic leukemia/lymphoma in a patient diagnosed during pregnancy
Introduction
Relapse following initial remission of adult acute lymphoblastic leukemia (ALL) occurs in approximately 50% of adult cases (1). In most patients the relapsed leukemic cells maintain the immunophenotypic and cytogenetic changes of the original leukemia, or obtain additional abnormalities, i.e., demonstrate clonal evolution. In rare cases the relapsed leukemia cells exhibit a new immunophenotype. Some of these cases have been labeled “lineage switch”, in which there is a loss of lineage defining markers of one lineage and gain of lineage defining markers of another lineage in the cells. In most reported cases of lineage switch patients with B-cell lymphoblastic leukemia/lymphoma (B-ALL) relapsed with acute myeloid or monocytic leukemia (AML) (2). Lineage switch is being reported more frequently following immunotherapy with either blinatumomab or CAR-T cells and is providing insight into the pathogenesis of acute leukemia (3-8). Currently we report a patient with T-cell lymphoblastic leukemia/lymphoma (T-ALL) who relapsed with AML 50 days after the initiation of standard T-ALL chemotherapy. When the patient relapsed a second time her cells demonstrated the T-ALL phenotype a second time.
Case presentation
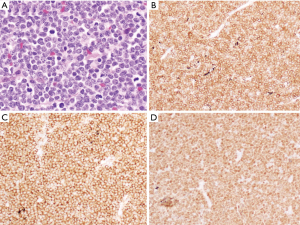
A 32-year-old Hispanic female presented at 30 weeks gestation with rapidly progressive lymph node swelling in the neck, axillae and inguinal regions. Her symptoms occurred over 2 weeks but were not associated with fever, night sweats or weight loss. On physical examination the patient had multiple cervical, axillary and inguinal lymph nodes up to 3 cm. There were no hepatosplenomegaly or skin rashes. The white blood cell count was 23,600×109/L with 64% neutrophils, 6% bands, 18% lymphocytes, 4% monocytes, 1% eosinophils, and 3% blasts. The hemoglobin was 10.3 gm/dL and the platelet count was 248,000×109/L. The LDH was 326 U/L. An inguinal lymph node biopsy showed effacement of the node with diffuse proliferation of atypical mononuclear cells (Figure 1). On immunohistochemistry the cells were positive for CD3, CD5, CD34, and TdT and negative for CD20, CD30, EBV (LMP-1) and CD1a (Table 1). Ki-67 was positive in >95% of neoplastic cells. Flow cytometry demonstrated an immature T cell population expressing CD2, CD3, CD5, CD7, CD8 (heterogeneous), CD56 (dim), CD1a (very dim), TCR alpha/beta and TdT (Table 1). The cells were negative for CD16, CD25, CD10 and TCR gamma/delta.
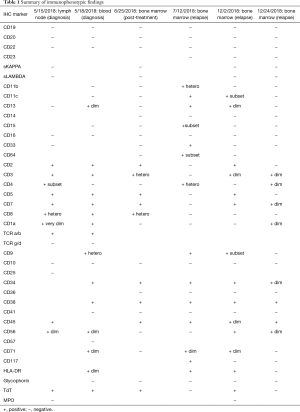
Full table
Due to the patient’s pregnancy a bone marrow was deferred. However peripheral blood flow cytometry demonstrated an immature T cell population (5% of total) expressing CD2, CD3, CD5, CD7, CD8, CD56 (dim), CD71 (dim), CD1a (dim), CD34, TCR alpha/beta and TdT. The cells were negative for CD4, CD16, CD57, CD10 and TCR gamma/delta. A Nextgen sequencing panel (Genoptix next course complete, 236 gene) demonstrated mutations in ALK, c.1277G>C; p.S426T, allelic frequency 48% and MAP3K14, c.428G>A; p.A143G, allelic frequency 53%. Cytogenetics were normal, 46,XX.
The patient was treated per the Children’s Oncology Group AALL0434 protocol with the exception that PEG-asparaginase and intrathecal methotrexate were withheld due to her pregnancy. She received prednisone 2 mg/kg daily on days 1−28, vincristine 2 mg on days 1, 8, 15, and 22, and daunorubicin 25 mg/m2 on days 1, 8, 15, and 22. The patient became preeclamptic and was induced on day 30 of therapy (34 weeks 6 days gestation). The baby weighed 2,590 grams and had Apgar scores of 8 and 8. The baby was discharged from the hospital after one week and is doing well.
Bone marrow aspiration and biopsy performed on day 34 showed 1% residual blasts expressing CD2, CD3 (heterogeneous), CD5, CD7, CD8 (heterogeneous), CD34, TdT, and CD38 but negative for CD4 and CD1a. The patient then received the “B cycle” of hyper-CVAD (methylprednisolone, high-dose methotrexate 1 gm/m2, and high dose cytarabine 3 gm/m2 every 12 hours for 4 doses) and was discharged to home. On day 12 the patient was readmitted with fever. The white count was 2,500×109/L with 4% neutrophils, 65% lymphocytes, 2% monocytes, 1% eosinophils, and 28% blasts. The hemoglobin was 9.4 gm/dL and the platelet count was 8,000×109/L. A repeat bone marrow showed 16% blasts that on flow cytometry expressed CD13, CD33, CD34, CD117, HLA-DR, CD11b (heterogeneous), CD11c, CD15 (subset), CD64 (subset), CD9 (heterogeneous), CD71 (dim), CD38, and CD4 (heterogeneous). The findings were consistent with AML (Figure 2). Nextgen sequencing again showed mutations in ALK, c.1277G>C; p.S426T, allelic frequency 51% and MAP3K14, c.428G>A; p.A143G, allelic frequency 56%. Cytogenetics were normal, 46,XX. After these results were obtained, the original lymph node specimen was assessed by immunohistochemistry for CD13, CD33 and myeloperoxidase. The latter results were negative.
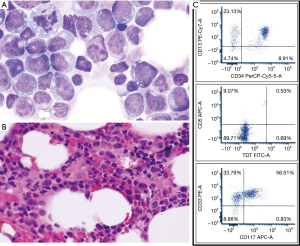
The patient then received cytarabine, 3 g/m2 iv over three hours once daily on days 1–5, mitoxantrone, 60 mg/m2 on day 2 and etoposide, 150 mg/m2 on days 1, 3 and 5. A post induction bone marrow showed no evidence of disease. The patient received 2 cycles of high dose cytarabine consolidation and underwent allogeneic stem cell transplantation from an HLA-matched sibling using busulfan, fludarabine and alemtuzumab as a conditioning regimen. The patient tolerated the transplant well, however a bone marrow done on day 30 demonstrated an immature T-cell population (62% of total) expressing CD3, CD2, CD5, CD7, CD34, TdT (dim), HLA-DR, CD56, CD38 and CD71 (dim) with aberrant expression of CD13 consistent with relapsed T-ALL. Next generation sequencing showed persistence of the two original mutations (ALK, c.1277G>C; p.S426T, allelic frequency 37%, and MAP3K14, c.428G>A; p.A143G, allelic frequency 53%), as well as a new mutation in ARID1A, c.5506_5513delGAGTTTGAinsTTTG; p.E1836Ffs*46, allelic frequency 36%. The patient also demonstrated a new FLT3 TKD mutation which was not present on the two prior samples. Cytogenetics demonstrated a new abnormality, 46,XX,del(6)(q13q21)/46,XX which has been described in T-cell lymphoblastic leukemia/lymphoma. The patient received several induction therapies, including clofarabine plus cytarabine, azacytidine plus venetoclax, alectinib, and prednisone plus vincristine without response. The patient expired 9 months after her original diagnosis.
Discussion
Rossi et al. studied the incidence of lineage switch in 1,482 pediatric patients with acute leukemia (2). Overall 9 cases (0.6%) of lineage switch occurred, seven from B-ALL to AML and two from AML to B-ALL. Switches occurred at a median of 15 days from initiation of therapy (range, 8 days to 6 months), with five switches occurring before day 15. Abnormalities in the 11q23/MLL gene were detected in seven cases. More recently, several cases of lineage switch from B-ALL to AML have been reported following CD19-targeted immunotherapies, including blinatumomab and CAR-T cells (3-8). In one case of B-ALL to AML on blinatumomab, the leukemia reverted back to B-ALL after the blinatumomab was discontinued (6). This has led investigators to propose that immune pressure exposes the inherent plasticity of the leukemic stem cell (7).
To date, cases of lineage switch from T-ALL to AML have only rarely been reported. Ittel et al. reported a case of T-ALL with t(10;11)(q22;q23)/MLL-TET1 who relapsed with AML 14 months after initial diagnosis (9). In this case the AML-TET1 fusion transcript was present in the AML cells at relapse even though the original TCR and IGH rearrangements were no longer present. Paganin described a patient with T-ALL and 46,XY,del(12)(p12) who relapsed with AML 1.7 years after diagnosis (10). This patient had different cytogenetic abnormalities at relapse raising the possibility of a therapy related AML rather than lineage switch. Higuchi et al. reported a case of T-ALL with t(6;11)(q27;q23) who upon 6th relapse demonstrated AML, although the cells maintained the MLL-MLLT4 fusion in the myeloid cells (11). Park et al. reported a case of lineage switch from B-ALL to T- ALL and then to AML. The AML occurred 45 days after therapy for the T-ALL (12).
There are several theories regarding the etiology of lineage switch (13-15). The most prominent is that both lineages occur from a common precursor cell. If the patient receives therapy that only targets leukemic cells more differentiated than the leukemic stem cell, the leukemic stem cell could then differentiate along a pathway that is not affected by the therapy. Other theories include dedifferentiation or cross-differentiation. Fujisaki et al. transplanted into SCID mice myeloid cells from a case in which T-ALL had undergone lineage switch to AML (14). Although the myeloid cells engrafted in the mice without cytokine administration, T-ALL developed in mice who received recombinant GM-CSF with the cell infusion. This study demonstrated that the differentiation of the leukemic stem cell can vary depending on the environment in which it is placed.
The current report is an unusual case in which a patient originally diagnosed with T-ALL relapsed rapidly with an AML phenotype. The leukemia reverted back to a T-cell phenotype in the second relapse. At initial diagnosis and in first relapse, the leukemic cells demonstrated normal cytogenetics and identical molecular mutations. In the second relapse the patient developed a new cytogenetic abnormality [del(6)(q13q21)] as well as a new mutation in ARID1A on next generation sequencing . In the absence of molecular testing of non-hematopoietic tissue, it is impossible to determine if the first two molecular findings were germ line or somatic. This case is further evidence of the plasticity of the leukemic stem cell.
Acknowledgments
None.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Informed Consent: Written informed consent was obtained from the patient for publication of this Case Report and any accompanying images.
References
- Jabbour E, O'Brien S, Konopleva M, et al. New insights into the pathophysiology and therapy of adult acute lymphoblastic leukemia. Cancer 2015;121:2517-28. [Crossref] [PubMed]
- Rossi JG, Bernasconi AR, Alonso CN, et al. Lineage switch in childhood acute leukemia: an unusual event with poor outcome. Am J Hematol 2012;87:890-7. [Crossref] [PubMed]
- Balducci E, Nivaggioni V, Boudjarane J, et al. Lineage switch from B acute lymphoblastic leukemia to acute monocytic leukemia with persistent t(4;11)(q21;q23) and cytogenetic evolution under CD19-targeted therapy. Ann Hematol 2017;96:1579-81. [Crossref] [PubMed]
- Haddox CL, Mangaonkar AA, Chen D, et al. Blinatumomab-induced lineage switch of B-ALL with t(4:11)(q21;q23) KMT2A/AFF1 into an aggressive AML: pre- and post-switch phenotypic, cytogenetic and molecular analysis. Blood Cancer J 2017;7:e607. [Crossref] [PubMed]
- Aldoss I, Song JY. Extramedullary relapse of KMT2A(MLL)-rearranged acute lymphoblastic leukemia with lineage switch following blinatumomab. Blood 2018;131:2507. [Crossref] [PubMed]
- Wölfl M, Rasche M, Eyrich M, et al. Spontaneous reversion of a lineage switch following an initial blinatumomab-induced ALL-to-AML switch in MLL-rearranged infant ALL. Blood Adv 2018;2:1382-5. [Crossref] [PubMed]
- Jacoby E, Nguyen SM, Fountaine TJ, et al. CD19 CAR immune pressure induces B-precursor acute lymphoblastic leukaemia lineage switch exposing inherent leukaemic plasticity. Nat Commun 2016;7:12320. [Crossref] [PubMed]
- Oberley MJ, Gaynon PS, Bhojwani D, et al. Myeloid lineage switch following chimeric antigen receptor T-cell therapy in a patient with TCF3-ZNF384 fusion-positive B-lymphoblastic leukemia. Pediatr Blood Cancer 2018;65:e27265. [Crossref] [PubMed]
- Ittel A, Jeandidier E, Helias C, et al. First description of the t(10;11)(q22;q23)/MLL-TET1 translocation in a T-cell lymphoblastic lymphoma, with subsequent lineage switch to acute myelomonocytic myeloid leukemia. Haematologica 2013;98:e166-8. [Crossref] [PubMed]
- Paganin M, Buldini B, Germano G, et al. A Case of T-cell Acute Lymphoblastic Leukemia Relapsed As Myeloid Acute Leukemia. Pediatr Blood Cancer 2016;63:1660-3. [Crossref] [PubMed]
- Higuchi Y, Tokunaga K, Watanabe Y, et al. Lineage switch with t(6;11)(q27;q23) from T-cell lymphoblastic lymphoma to acute monoblastic leukemia at relapse. Cancer Genet 2016;209:267-71. [Crossref] [PubMed]
- Park M, Koh KN, Kim BE, et al. Lineage switch at relapse of childhood acute leukemia: a report of four cases. J Korean Med Sci 2011;26:829-31. [Crossref] [PubMed]
- Hu T, Murdaugh R, Nakada D. Transcriptional and Microenvironmental Regulation of Lineage Ambiguity in Leukemia. Front Oncol 2017;7:268. [Crossref] [PubMed]
- Fujisaki H, Hara J, Takai K, et al. Lineage switch in childhood leukemia with monosomy 7 and reverse of lineage switch in severe combined immunodeficient mice. Exp Hematol 1999;27:826-33. [Crossref] [PubMed]
- Dorantes-Acosta E, Pelayo R. Lineage switching in acute leukemias: a consequence of stem cell plasticity? Bone Marrow Res 2012;2012:406796. [Crossref] [PubMed]
Cite this article as: Aujla A, Hanmantgad M, Islam H, Shakil F, Liu D, Seiter K. Lineage switch from T-cell lymphoblastic leukemia/lymphoma to acute myeloid leukemia and back to T-cell lymphoblastic leukemia/lymphoma in a patient diagnosed during pregnancy. Stem Cell Investig 2019;6:12.


