
Lineage choice decisions in B-cell development and leukemia
Every tissue in the body is founded by a particular type of cell that is termed stem cell. These cells are generally presumed to be tissue-specific and, as such, are also responsible for maintenance of the tissue throughout the lifespan of an organism. There are two fundamental aspects to the regular behaviour of tissues during their genesis and thereafter. The offspring of stem cells divide in a manner that is controlled to the social benefit of the organism to generate/ensure the bulk of tissue that is required. These dividing cells mature: they acquire individual characteristics to enable them to perform a unique function within the tissue. Within cancer cells aspects of this normal physiology are distorted. This means the many strands to understanding of the behaviour of normal cells cannot be loosened from the strands of efforts to unravel what goes wrong in cancer.
For many years, the blood cell system has provided a model system that has been used by many researchers to investigate how a stem cell can give rise to a wide variety of mature cell types. The principles that emerged to developmental biology have been applied to the structure of tissues throughout the body. However, many of the principles have been challenged by findings over a number of years. This has led to revision to the way we view blood cell development. In turn, this has impacted on our understanding of the origin and nature of leukemias, and cancer in general. “Classic” bifurcating tree maps for the development of the various blood and immune cells depict a single route to each of the mature cell types and strict compartments to haematopoietic stem cells (HSCs) and haematopoietic progenitor cells (HPCs). This model led to the notion that decision-making occurs much earlier than previously thought and within HSCs. However, developing HSCs and HPCs are much more versatile than described in the classic model. To directly address the versatility of HSCs and HPCs, Zhang et al. [2018] designed an initial screen to identify candidate transcription factors mainly expressed in HSCs and HPCs but not in lineage-committed cells (1). Within the 15 transcription factors identified, only forced expression of Hoxb5 factor was able to reprogram committed progenitors of B cells into distinct subsets of T lymphocytes. The authors performed then an elegant series of experiments examining the basis for the selective regulation of Hoxb5-mediated B-cell reprogramming potential. The authors showed that the constitutive expression of Hoxb5 in all hematopoietic cells, including T lymphocytes, had a minimal effect on hematopoiesis. However, transient expression of Hoxb5 in precursor B cells was sufficient for stable conversion of B cells into T cells in vivo, and sustained expression of Hoxb5 was dispensable after the target cells committed to lineage conversion. These results convincingly showed that forced expression of Hoxb5 in precursor (pro/pre) B cells was sufficient to convert mouse B cells into T cells in vivo. However, the efficiency of Hoxb5-induced B cell-to-T cell conversion was very low, indicating that a minority of precursor B cells overexpressing Hoxb5 were reprogrammed into T cells in vivo, but the majority were able to differentiate normally into B cells. This cell-fate conversion was the consequence of Hoxb5-mediated repression of genes encoding B cell master regulators, activation of genes encoding T cell regulators and regulation of genes encoding chromatin and epigenetic modifiers and remodelers. Thus, they further found that Hoxb5 directly targeted Ebf1, Pax5, Bcl11a, Foxp1 and Foxo1, consistent with published reports that repression of Pax5 and Ebf1 is crucial for the B cell-to-T cell conversion (2,3). Likewise, Hoxb5 directly targeted transcription factors with important roles in the regulation of T cell fate as Lmo2, Nfatc1, Tcf12, Prdm1 and Runx2.
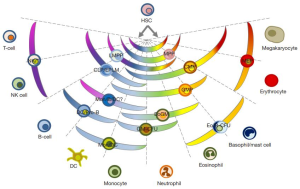
The results are both attractive and provocative in equal measure. On one hand, the conversion of precursor B-cells into T cells indicates that, after having selected a cell lineage, they can still “step sideways” to adopt alternative, closely related, fates. Thus, these findings are a clear support for the pair-wise model of hematopoiesis proposed by Brown and Ceredig (4-6). Notably, the pair-wise model does not prescribe a single route from HSCs to each of the end cell types. Instead, the model envisages a spectrum of options arranged in a particular manner (Figure 1). According to this model, differentiation options are envisaged as a series of invariant pair-wise developmental relationships with cells becoming gradually biased from the hematopoietic stem cell stage of development towards producing one cell type or another. This major change to our understanding of haematopoiesis is supported by the in vivo conversion of precursor B-cells into T cells and impacts on how we view leukemia development (Figure 2).
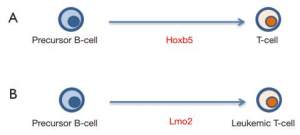
During leukemogenesis a normal cell acquires a new but inappropriate (malignant) identity to give rise to a clonal aberrant population. This is only possible if the oncogenic event initiating cancer had an inherent reprogramming capacity, to be able to lead to the change in cellular identity (7,8). LMO2 is one of the most frequent drivers of childhood T-cell acute lymphoblastic leukemia (T-ALL) (9) and has been identified as one of the transcription factors regulated by Hoxb5 during the in vivo conversion of precursor B-cells into T cells (1). Likewise, recent findings have shown that LMO2 functions as a “hit-and-run” oncogene that acts at an early stage of T-cell leukemogenesis to reprogram hematopoietic stem/progenitor cells (HS/PCs) for T-cell malignancy, showing that activity of the Lmo2 oncogene restricted to HS/PCs can induce malignancies in mice that are of a T-cell stage of differentiation (10,11). However, the permissiveness for development of T-ALL seems to be associated with wider windows of differentiation than previously appreciated. Restricted Cre-mediated activation of Lmo2 at different stages of B-cell development, including pro-pre B cells, induced systematically and unexpectedly T-ALL that closely resembled those of their natural counterparts (10). Biological barriers exist to prevent cells from changing their identity in this manner in order to avoid the risk of malignant transformation (7,8,12,13). In B-cell malignancies it has been shown that loss of p53 is a frequent occurrence and facilitates the pathological reprogramming to a malignant B-cell phenotype (14,15). Similarly, a significant proportion of T-ALL in all our murine models carried p53 loss-of-function mutations facilitating pathological reprogramming to a malignant T-cell phenotype (10). It will be worthy to see if p53 is also a biological barrier for Hoxb5-mediated B-cell reprogramming potential (Figure 2A). These findings led us to propose that T-ALL is the result of an inappropriate lineage-decision making process occurring via a reprogramming-like mechanism which can even start within a B-cell (Figure 2B). Nonetheless, the observation that transient expression of Hoxb5 was sufficient for stable conversion of precursor B cells into T cells in vivo raises a potential new avenue for the full understanding of hematopoietic development and the genesis of leukemia.
Acknowledgements
We are indebted to all members of our groups for useful discussions and for their critical reading of the manuscript. Research in C Vicente-Dueñas group is partially supported by FEDER, “Miguel Servet” Grant (CP14/00082 - AES 2013-2016) from the Instituto de Salud Carlos III (Ministerio de Economía y Competitividad), “Fondo de Investigaciones Sanitarias/Instituto de Salud Carlos III” (PI17/00167). Research in I Sánchez-García (ISG) group is partially supported by FEDER and by MINECO (SAF2012-32810, SAF2015-64420-R and Red de Excelencia Consolider OncoBIO SAF2014-57791-REDC), by Junta de Castilla y León (UIC-017, and CSI001U16). ISG lab is a member of the EuroSyStem and the DECIDE Network funded by the European Union under the FP7 program. ISG has been supported by the German Carreras Foundation (DJCLS R13/26). G Rodríguez-Hernández was supported by FSE-Conserjería de Educación de la Junta de Castilla y León (CSI001-15).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Zhang M, Dong Y, Hu F, et al. Transcription factor Hoxb5 reprograms B cells into func-tional T lymphocytes. Nat Immunol 2018;19:279-90. Erratum in: Publisher Correction: Transcription factor Hoxb5 reprograms B cells into functional T lymphocytes. [Nat Im-munol 2018]. [Crossref] [PubMed]
- Souabni A, Cobaleda C, Schebesta M, et al. Pax5 promotes B lymphopoiesis and blocks T cell development by repressing Notch1. Immunity 2002;17:781-93. [Crossref] [PubMed]
- Nechanitzky R, Akbas D, Scherer S, et al. Transcription factor EBF1 is essential for the maintenance of B cell identity and prevention of alternative fates in committed cells. Nat Immunol 2013;14:867-75. [Crossref] [PubMed]
- Ceredig R, Rolink AG, Brown G. Models of haematopoiesis: seeing the wood for the trees. Nat Rev Immunol 2009;9:293-300. [Crossref] [PubMed]
- Brown G, Ceredig R, Tsapogas P. The Making of Hematopoiesis: Developmental Ancestry and Environmental Nurture. Int J Mol Sci 2018;19. [Crossref] [PubMed]
- Brown G, Sanchez-Garcia I. Diversity, Versatility and Leukaemia. New York: Nova Science Publishers Inc., 2016.
- Vicente-Dueñas C, Hauer J, Cobaleda C, Borkhardt A, Sánchez-García I. Epigenetic Priming in Cancer Initiation. Trends Cancer 2018;4:408-17. [Crossref] [PubMed]
- González-Herrero I, Rodríguez-Hernández G, Luengas-Martínez A, et al. The Making of Leukemia. Int J Mol Sci 2018;19. [Crossref] [PubMed]
- Liu Y, Easton J, Shao Y, et al. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat Genet 2017;49:1211-8. [Crossref] [PubMed]
- García-Ramírez I, Bhatia S, Rodríguez-Hernández G, et al. Lmo2 expression defines tumor cell identity during T-cell leukemogenesis. EMBO J 2018;37. [Crossref] [PubMed]
- Rodríguez-Hernández G, Bhatia S, Vicente-Dueñas C, et al. T-cell leukemogenesis is an inappropriate lineage decision-making process: implications for precision oncology. Mol Cell Oncol 2018;5:e1497860. [Crossref] [PubMed]
- Rodríguez-Hernández G, Hauer J, Martín-Lorenzo A, et al. Infection Exposure Promotes ETV6-RUNX1 Precursor B-cell Leukemia via Impaired H3K4 Demethylases. Cancer Res 2017;77:4365-77. [Crossref] [PubMed]
- Martín-Lorenzo A, Auer F, Chan LN, et al. Loss of Pax5 Exploits Sca1-BCR-ABLp190 Susceptibility to Confer the Metabolic Shift Essential for pB-ALL. Cancer Res 2018;78:2669-79. [Crossref] [PubMed]
- Vicente-Dueñas C, Fontán L, Gonzalez-Herrero I, et al. Expression of MALT1 oncogene in hematopoietic stem/progenitor cells recapitulates the pathogenesis of human lymphoma in mice. Proc Natl Acad Sci U S A 2012;109:10534-9. [Crossref] [PubMed]
- Vicente-Dueñas C, Romero-Camarero I, González-Herrero I, et al. A novel molecular mechanism involved in multiple myeloma development revealed by targeting MafB to haematopoietic progenitors. EMBO J 2012;31:3704-17. [Crossref] [PubMed]
Cite this article as: Raboso-Gallego J, Casado-García A, Rodríguez-Hernández G, Vicente-Dueñas C, Sánchez-García I. Lineage choice decisions in B-cell development and leukemia. Stem Cell Investig 2018;5:46.


