Pharmacological inhibition of aberrant transcription factor complexes in inversion 16 acute myeloid leukemia
Within the hematopoietic system, core binding factor beta (CBFβ) normally forms a complex with the master hematopoietic regulator, RUNX1 (1) and stabilizes its binding to DNA (2). In core binding factor (CBF) acute myeloid leukemias (AMLs), chromosomal rearrangements alter either RUNX1 or CBFβ, thus dysregulating normal hematopoiesis. These AMLs are classified as having a favourable prognosis with complete remission expected following cytarabine and anthracycline based chemotherapy (3). However, relapse rates remain high and durable responses to salvage chemotherapy remain uncommon, therefore there is still an urgent need for new therapies.
The inversion 16 (inv(16)) CBF AML subtype does not involve the RUNX1 gene directly, but arises as a consequence of the expression of an abnormal CBFβ-SMMHC (core binding factor beta-heavy chain of smooth muscle myosin) fusion protein (4) causing a block in hematopoietic differentiation (5). CBFβ-SMMHC can act in a dominant negative manner to CBFβ and interacts with RUNX1 with 10 fold higher affinity due to the presence of the High Affinity Binding Domain at the N-terminus of SMMHC. It was therefore originally thought that CBFβ-SMMHC simply sequesters RUNX1 and stops its binding to DNA. However, this was refuted by an inv(16) knock-in mouse model which showed that RUNX1 activity was crucial to leukemogenesis as knockout of RUNX1 prevented the expected CBFβ-SMMHC mediated differentiation block (6).
Initial insights into the action of CBFβ-SMMHC came from a genome-wide analysis of its binding sites in an inv(16) cell line and a patient sample, which found that RUNX1 appeared to be co-localizing with the fusion protein at its target genes (7). After CBFβ-SMMHC knockdown by shRNA, cells were found to down-regulate genes associated with a stem cell phenotype and self-renewal and up-regulate more differentiated myeloid cell related genes, indicating that the fusion protein was directly responsible for the differentiation block. Pulikkan and colleagues (8) have taken this work further by exploring the mechanisms of growth deregulation in this type of AML by pharmacologically inhibiting the CBFβ-SMMHC:RUNX1 complex directly.
The inhibition of transcription factors has long been thought to be a holy grail in therapeutics as these molecules were considered unsuitable for drug targeting. The few exceptions include natural ligands, such as All-Trans Retinoic Acid (ATRA) in Acute Promyelocytic Leukemia which was a first example of a small molecule compound targeting an aberrant transcription factor, PML-RARA. The authors of the Pulikkan et al. study, from the groups of Lucio Castilla and John Bushweller (8), had previously identified inhibitors from a pharmacological screen using a fluorescence resonance energy transfer (FRET) assay based on blocking the interaction between RUNX1 and CBFβ-SMMHC. Using further modifications they made the compound highly selective for CBFβ-SMMHC:RUNX1, with the most effective molecule being AI.10.49 (9). They showed that inv(16) AML had marked overexpression of MYC and that after treatment with their compound, MYC was one of the most down-regulated genes. Furthermore, knockdown of MYC using RNA interference conferred a significant survival advantage in xenotransplantation experiments and this recapitulated the phenotypic effects of AI.10.49. However, the mechanism by which MYC is overexpressed in inv(16) AML was previously not known.
MYC is well known to be overexpressed across many types of AML (10) and overexpressing MYC in normal hematopoietic progenitors led to the development of AML in mouse models (11). The mechanism of how MYC is overexpressed is understood in NPM1 mutated AML; here NPM1c inhibits members of the E3 ligase family that normally promote the proteasomal degradation of the MYC protein (12). In addition, CEBPA normally negatively regulates MYC via an E2F binding site in the MYC promoter and consequently in CEBPA mutated AML, this repression is relieved (13). Finally, secondary activating mutations in signaling receptors such as FLT3-ITD and cKIT can upregulate MYC by STAT5 signaling (14). Inv(16) deploys an altogether different mechanism.
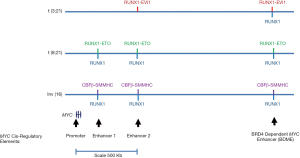
Pulikkan and colleagues (8) showed by chromatin immunoprecipitation (ChIP) assays that RUNX1 normally binds to 3 downstream MYC enhancer elements and that use of their inhibitor increases RUNX1 binding at this position, as well as globally. Furthermore, they showed by a chromatin conformation capture assay that these three enhancer elements directly interact with the MYC promoter and each other and that treatment with the compound strengthened these interactions. Functionally, they showed that deletion of any of these enhancers by CRISPR-Cas9 gene editing reduced MYC transcription and cell viability. The most distal of these three enhancers has been previously characterised as a BRD4-mediated MYC enhancer (BDME) (15) (Figure 1). In defining the molecular details of how the compound interferes with aberrant MYC regulation, Pulikkan et al. (8) showed that BRD4 knockdown by RNA interference in the inv(16) cell line reduced MYC transcripts levels. They went on to show synergism between the BRD4 inhibitor JQ1 (16) and their inhibitor AI.10.49 in vitro whereby cell viability was decreased and in vivo whereby xenotransplanted mice survived for much longer.
These results raise exciting therapeutic prospects as the combination of JQ1 and AI.10.49 could have a role in the relapsed/refractory setting or frontline in those patients too frail for intensive chemotherapy. However, there will need to be several further developments before such drugs may be suitable for therapeutic use. The half-life of AI.10.49 is 380 minutes in mice (9) and needs to be increased to be suitable for humans. The half-life of JQ1 is also very short at around 1 hour (16), but there have been several attempts to produce modified BET inhibitors; OTX001 looks like one of the most promising with a half-life of 6 hours in humans, making it suitable for oral dosing four times a day, and with several clinical responses reported in case series (17).
Another important question relates to whether this therapy will work on quiescent pre-leukemic and leukemic stem cells where MYC expression may be low (18). It is increasingly recognized that such cells may survive intensive chemotherapy and may act as a reservoir of cells for relapse (19). Whilst it is thought that pre-leukemic and leukemic stem cells are aberrantly regulated by the CBFβ-SMMHC fusion protein as the inv(16) translocation is a first hit mutation in AML, it remains to be seen whether the presence of the fusion protein is sufficient to de-repress MYC transcription.
Shi and colleagues (15) showed that in a MLL-AF9 AML context, BRG1, an ATPase member of the SWI/SNF complex was key to establish and open chromatin conformation at the BDME enhancer. In inv(16) AML, BRG1 binds not only to this enhancer but also two other RUNX1-binding enhancers and the MYC promoter and use of AI.10.49 reduces BRG1 to all of these cis-regulatory elements (8). Furthermore, during hematopoietic differentiation RUNX1 has previously been described as acting as a repressor by recruiting RING1B, a member of the polycomb-repressive complex (PRC) (20). RING1B binding to each of the three downstream MYC enhancers was shown to be increased after AI.10.49 treatment and in a time course experiment following the addition of AI.10.49, it was demonstrated that as BRG1 binding decreased, RING1B binding increased (8). Consequently there appears to be competition between active transcriptional complexes including BRG1 and the gene silencing machinery including RING1B and the use of AI.10.49 pushes this equilibrium towards repression.
The finding that RUNX1 replaces BRG1 at active enhancers with RING1B, a member of the PRC, confirms multiple reports showing that RUNX1 can act as a transcriptional repressor in both normal and abnormal hematopoiesis (1,21). In our work we also found an interplay between RUNX1 and another CBF fusion protein, RUNX1-ETO, the product of the t(8;21) translocation. RUNX1 dynamically binds to a crucial cis-regulatory element of the Cyclin D2 (CCND2) gene and inhibits its transcription, thus regulating the cell cycle at the G1/S checkpoint (22). In t(8;21) AML, the RUNX1-ETO fusion protein competes with RUNX1 for binding an upstream element and cooperates with the AP-1 transcription factor family in binding to the CCND2 promoter, thus leading to a de-repression of transcription. In the context of the MYC enhancers, it would be very interesting to know which other proteins are present at the active MYC enhancers which cooperate to drive MYC transcription. Mandoli and colleagues showed that globally at CBFβ-SMMHC:RUNX1 binding sites that GATA and ETS factors such as FLI1, PU.1 and ERG bind (7) but the full composition of the factor complex at MYC enhancers is unknown.
Fusion protein interference with RUNX1-mediated MYC regulation may also play also a role in other well-characterised types of CBF AML, such as the t(8;21) and the t(3;21) which fuses the RUNX1 DNA-binding domain to the EVI1 transcriptional repressor. Figure 1 shows the binding locations of these complexes as well as of RUNX1 to MYC cis-regulatory elements based upon ChIP-seq data from the Mandoli and colleagues study (7) and our own published work (23,24). Consequently, when RUNX1-ETO was depleted by the use of siRNA, we found a decrease in MYC transcript levels.
The inhibition of protein complex formation provides an intriguing way of treating cancer and requires a significant structural and biological understanding of the proteins involved. Another inhibitor from the Bushweller lab allosterically alters the binding of key CBFβ residues to RUNX1 and hence decreases RUNX1 binding at its target sites and could be used to exploit the dependency of CBF AML on wild-type RUNX1 (25). This inhibitor had efficacy on a variety of AML cell lines including Kasumi-1, which is driven by the t(8;21) translocation but remarkably had minimal effect on normal hematopoiesis. Another example where such a strategy may be employed is in NPM1 mutated AML, which is the most common driver mutation in karyotypically normal AML. NPM1 is required for the cytoplasmic localization and destabilization of Fbw7γ in order to prevent nuclear degradation of several crucial AML oncoproteins such as MYC, NOTCH, CYCLIN E and JUN (12). A recurrent protein-interacting domain in NPM1 has been identified which could be therapeutically targeted (26). Furthermore, in inv(16) AML, there is a 28 amino acid Assembly Competence Domain near the C-terminus which is involved in the oligomerization of the CBFβ-SMMHC protein and this interface could potentially be targeted to prevent the formation of oligomeric complexes. Such a strategy has already been carried out in t(8;21) AML where a polypeptide (NC128) was used to inhibit oligomerization of RUNX1-ETO via the Nervy Homology Region 2 (27).
One further major approach to therapeutics has been the coupling of molecules which recruit an E3 ubiquitin ligase to small molecules which can bind a transcription factor. This allows for the ubiquitination and subsequent destruction of transcription factors by the proteasome and such molecules are known as proteolysis targeting chimaeras (PROTACs) (28).
Currently we still treat AML with chemotherapy, often with treatment regimes that have not changed for decades. The experiments described above suggest that we may be at the advent of developing methods to beat cancers at the game of reprogramming transcriptional networks by the development of truly novel and potent therapies for patients.
Acknowledgements
S Potluri is funded by a Medical Research Council and Leuka Clinical Fellowship. Leukemia work in the Bonifer Lab including Daniel Coleman’s position is funded by a Bloodwise Programme Grant.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Bonifer C, Levantini E, Kouskoff V, et al. Runx1 Structure and Function in Blood Cell Development. Adv Exp Med Biol 2017;962:65-81. [Crossref] [PubMed]
- Tahirov TH, Inoue-Bungo T, Morii H, et al. Structural analyses of DNA recognition by the AML1/Runx-1 Runt domain and its allosteric control by CBFbeta. Cell 2001;104:755-67. [Crossref] [PubMed]
- Grimwade D, Walker H, Oliver F, et al. The importance of diagnostic cytogenetics on outcome in AML: analysis of 1,612 patients entered into the MRC AML 10 trial. The Medical Research Council Adult and Children's Leukaemia Working Parties. Blood 1998;92:2322-33. [PubMed]
- Liu P, Tarlé SA, Hajra A, et al. Fusion between transcription factor CBF beta/PEBP2 beta and a myosin heavy chain in acute myeloid leukemia. Science 1993;261:1041-4. [Crossref] [PubMed]
- Xue L, Pulikkan JA, Valk PJ, et al. NrasG12D oncoprotein inhibits apoptosis of preleukemic cells expressing Cbfβ-SMMHC via activation of MEK/ERK axis. Blood 2014;124:426-36. [Crossref] [PubMed]
- Hyde RK, Zhao L, Alemu L, et al. Runx1 is required for hematopoietic defects and leukemogenesis in Cbfb-MYH11 knock-in mice. Leukemia 2015;29:1771-8. [Crossref] [PubMed]
- Mandoli A, Singh AA, Jansen PW, et al. CBFB-MYH11/RUNX1 together with a compendium of hematopoietic regulators, chromatin modifiers and basal transcription factors occupies self-renewal genes in inv(16) acute myeloid leukemia. Leukemia 2014;28:770-8. [Crossref] [PubMed]
- Pulikkan JA, Hegde M, Ahmad HM, et al. CBFβ-SMMHC Inhibition Triggers Apoptosis by Disrupting MYC Chromatin Dynamics in Acute Myeloid Leukemia. Cell 2018;174:172-186.e21. [Crossref] [PubMed]
- Illendula A, Pulikkan JA, Zong H, et al. Chemical biology. A small-molecule inhibitor of the aberrant transcription factor CBFβ-SMMHC delays leukemia in mice. Science 2015;347:779-84. [Crossref] [PubMed]
- Slovak ML, Ho JP, Pettenati MJ, et al. Localization of amplified MYC gene sequences to double minute chromosomes in acute myelogenous leukemia. Genes Chromosomes Cancer 1994;9:62-7. [Crossref] [PubMed]
- Luo H, Li Q, O'Neal J, et al. c-Myc rapidly induces acute myeloid leukemia in mice without evidence of lymphoma-associated antiapoptotic mutations. Blood 2005;106:2452-61. [Crossref] [PubMed]
- Bonetti P, Davoli T, Sironi C, et al. Nucleophosmin and its AML-associated mutant regulate c-Myc turnover through Fbw7 gamma. J Cell Biol 2008;182:19-26. [Crossref] [PubMed]
- Johansen LM, Iwama A, Lodie TA, et al. c-Myc is a critical target for c/EBPalpha in granulopoiesis. Mol Cell Biol 2001;21:3789-806. [Crossref] [PubMed]
- Chatterjee A, Ghosh J, Ramdas B, et al. Regulation of Stat5 by FAK and PAK1 in Oncogenic FLT3- and KIT-Driven Leukemogenesis. Cell Rep 2014;9:1333-48. [Crossref] [PubMed]
- Shi J, Whyte WA, Zepeda-Mendoza CJ, et al. Role of SWI/SNF in acute leukemia maintenance and enhancer-mediated Myc regulation. Genes Dev 2013;27:2648-62. [Crossref] [PubMed]
- Filippakopoulos P, Qi J, Picaud S, et al. Selective inhibition of BET bromodomains. Nature 2010;468:1067-73. [Crossref] [PubMed]
- Doroshow DB, Eder JP, LoRusso PM. BET inhibitors: a novel epigenetic approach. Ann Oncol 2017;28:1776-87. [Crossref] [PubMed]
- Scognamiglio R, Cabezas-Wallscheid N, Thier MC, et al. Myc Depletion Induces a Pluripotent Dormant State Mimicking Diapause. Cell 2016;164:668-80. [Crossref] [PubMed]
- Ford AM, Mansur MB, Furness CL, et al. Protracted dormancy of pre-leukemic stem cells. Leukemia 2015;29:2202-7. [Crossref] [PubMed]
- Yu M, Mazor T, Huang H, et al. Direct recruitment of polycomb repressive complex 1 to chromatin by core binding transcription factors. Mol Cell 2012;45:330-43. [Crossref] [PubMed]
- Ter Elst A, Ma B, Scherpen FJ, et al. Repression of vascular endothelial growth factor expression by the runt-related transcription factor 1 in acute myeloid leukemia. Cancer Res 2011;71:2761-71. [Crossref] [PubMed]
- Martinez-Soria N, McKenzie L, Draper J, et al. The Oncogenic Transcription Factor RUNX1/ETO Corrupts Cell Cycle Regulation to Drive Leukemic Transformation. Cancer Cell 2018. In Press.
- Loke J, Assi SA, Imperato MR, et al. RUNX1-ETO and RUNX1-EVI1 Differentially Reprogram the Chromatin Landscape in t(8;21) and t(3;21) AML. Cell Rep 2017;19:1654-68. [Crossref] [PubMed]
- Ptasinska A, Assi SA, Martinez-Soria N, et al. Identification of a dynamic core transcriptional network in t(8;21) AML that regulates differentiation block and self-renewal. Cell Rep 2014;8:1974-88. [Crossref] [PubMed]
- Illendula A, Gilmour J, Grembecka J, et al. Small Molecule Inhibitor of CBFβ-RUNX Binding for RUNX Transcription Factor Driven Cancers. EBioMedicine 2016;8:117-31. [Crossref] [PubMed]
- Di Matteo A, Franceschini M, Paiardini A, et al. Structural investigation of nucleophosmin interaction with the tumor suppressor Fbw7γ. Oncogenesis 2017;6. [Crossref] [PubMed]
- Wichmann C, Chen L, Heinrich M, et al. Targeting the oligomerization domain of ETO interferes with RUNX1/ETO oncogenic activity in t(8;21)-positive leukemic cells. Cancer Res 2007;67:2280-9. [Crossref] [PubMed]
- Winter GE, Buckley DL, Paulk J, et al. DRUG DEVELOPMENT. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science 2015;348:1376-81. [Crossref] [PubMed]
Cite this article as: Potluri S, Coleman D, Bonifer C. Pharmacological inhibition of aberrant transcription factor complexes in inversion 16 acute myeloid leukemia. Stem Cell Investig 2018;5:30.


