The ved protein patterning in zebrafish embryos
Introduction
The vertebrate vent-family transcription factors play a crucial role in the establishment of the dorsoventral axis in embryos, by repressing the organizer genes. Three zebrafish homeobox genes, vox/vega1, vent/vega2 and ved encode transcriptional repressors implicated in the dorsoventral patterning (1-7). Some organizer genes such as bozozok/dharma, goosecoid, and chordin repress the expression of vent-family genes in dorsal region. A transcriptional repressor Dha/Boz acts due to the Eh1 domain—the conserved EFDQYLN peptide located in the C-terminal region (7). Vent-family genes also have an important function in down-regulation in ventral endoderm development and the separation of endoderm from mesoderm and ectoderm (8). Vox, vent and ved are expressed in overlapping domains, consistent with a cell-autonomous interaction, potentially by physical protein-protein cooperation with cas or pou. Moreover, they influence a differentiation of some ectoderm and mesoderm derivatives. Recently it was shown in medaka fish embryos (9), that vox was expressed in the developing neural tube similarly to central nervous system (CNS) markers. Thus the vent family genes, in addition to their fundamental role in body axis formation, may be involved in regionalization of vertebrate CNS.
There is a very interesting work on the Integrator Complex Subunit 6 (Ints6) (10). The only reported role for the Ints6 was to mediate a processing of snRNA of the spliceosome. The authors had isolated a recessive dorsalizing maternal-effect mutation disrupting the Ints6 gene. The embryos from mutant mothers failed to maintain the expression of BMP ligands, lacked vox and ved full expression after sphere stage, displayed delayed cell movements during gastrulation, and were severely dorsalized. Thus, Ints6 restricts the organizer to dorsal domains, preventing it from extending around the margin into ventral domains. A visible vanishing of the vox and ved mRNA occurring after mid-blastula transition indicates that the maternally stored mRNA degraded, and their synthesis de novo on the zygotic genome is blocked. Whether a processing of the vox and ved mRNA is confined, hybridization in situ should reveal the incompletely processed templates. Thus in the mutants there takes place a disturbance in processing of mRNA for some booster of the vent-genes activity on the zygotic genome. These data demonstrate that the gene expression regulation is multilevel and may occur at the level of mRNA maturation. Furthermore, the mRNA for differentiation governors can be stored in inactive form, and only the activation of translation may let the protein be synthesized and reveal its regulatory function (11). Concerning the vent-family genes we have previously shown (12,13) that Xvent-2 mRNA was not active in tails of Xenopus embryos at the early tail bud stage. Although the Xvent-2 mRNA in Xenopus and vox mRNA in zebrafish embryos were detected in the tips of the tails, the Xvent-2 and vox proteins were revealed in other regions of the tails - along axial structures and around somites. We proposed that when the translation of the masked Xvent-2 (or vox) mRNA was activated in a tail bud cell, this transcription factor changed activity of target genes. As a result, the cell came to differentiation and migrated to other space. That was why mRNA and the encoded protein were revealed at different regions of the embryo. These data coincided with the fact (14) that Vent2 was co-expressed with Snail2, a marker of neural crest cells, at late gastrula/neurula stages when neural crest cells retained their full developmental potential, but was down-regulated as these cells began to migrate and lost multipotency. According to our results (12,13), at tail bud stages the vent-family mRNAs were synthesized in pluripotent tail bud cells, but the proteins were revealed in derivative cells, migrating in the neural crest zone.
While the protein particularly is responsible for the function, it is necessary to pay more attention to the protein patterning. In this study we obtained antibodies against the ved protein, tested their specificity by Western-blot analysis and performed the whole mount immunostaining of zebrafish embryos. It was shown that the ved spatial-temporal pattern coincided with those of vox and vent obtained by using the appropriate antibodies (12,13).
Methods
Embryos, stages
All the Zebrafish embryos (MZspg793 and WT) were kindly donated by Dr. D. Onichtchouk (Albert-Ludwigs-University, Freiburg, Germany). About 10–30 embryos were taken for each test sample and about 5–10 for each control sample. Zebrafish embryos developmental stages were identified according to (15).
Production of antibodies
Rabbit polyclonal antibodies against Zebrafish ved and vox were produced by Almabion Company (Russia) using rvfkrnaylgaqdkaelc (for ved) and pvldvqepekktrphvpc (for vox) peptides. Then the antibodies were purified by affinity chromatography and conjugated with horseradish peroxidase. The same conjugate with rabbit polyclonal antibodies against human immunoglobulin (Imtek, Russia) was used in control experiments.
Western-blot analysis
The dechorionized embryos were homogenized in sample buffer and analyzed in Laemmli PAAG electrophoresis. Western-blot analysis was performed with single appropriate HRP-conjugated antibodies using SuperSignal Western Blot Enhancer Kit (Thermo Scientific, USA) according to the manufacturer recommendations.
Whole-mount immunostaining of embryos
Embryos were primarily fixed in MEMFA solution. Following procedures were described in (16).
Results and discussion
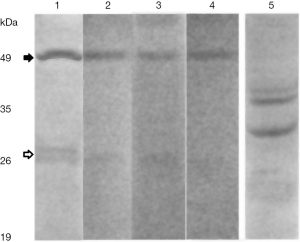
The ved protein was predicted to consist of 278 amino acids. The antibodies were arisen against the 18-amino acids peptide showing no homology with the other proteins. The Western-blot revealed (Figure 1) that the ved moved as one band higher than 50 kDa, and a duplet higher than 25 kDa. The heavy band was probably a dimer. The results of Western-blot were similar for the embryos at different stages of development and MZspg mutants. The vox showed quite good two bands. Such duplets were also found for some other proteins (17), probably due to different extent of phosphorylation. It seems really possible for the both vox and ved aminoacid sequences depict plenty of prospective phosphorylation sites. The Figure 1 also demonstrates an absence of cross-reaction between antibodies against vox and ved.
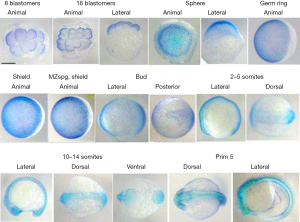
The whole-mount immunostaining revealed the ved protein in the embryos at the stages of 8–16 blastomeres and later on (Figure 2). It seems that ved protein was prestored maternally. Up to the shield stage the ved protein staining progressively covered the embryos except for dorsal side. The same staining was in maternal-zygotic mutant in pou5f3 gene (MZspg). It shows independence of the ved synthesis on the pou5f3. At the beginning of somitogenesis and at the pharyngula period stages ved was clearly seen alongside the neural tube, where the neural crest cells resided. Additionally at these stages the ved protein staining was revealed in the head region of the embryos, in presumptive eyes and was absent from the tail tips.
The staining with the appropriate antibodies could not reveal any differences in spatial-temporal distributions of the ved and the other vent-family proteins (12,13). Figure 3 demonstrates, for the comparison, the immunostaining of embryos with anti-vox and anti-human immunoglobulin antibodies. One can see that there is no difference in spatial distributions of the vox and ved proteins. The anti-human immunoglobulin antibodies do not interact with the embryos by any means.
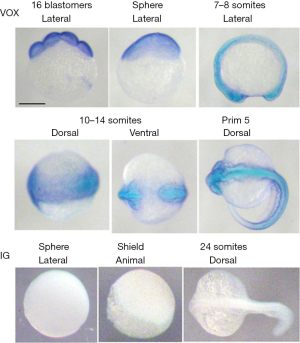
Thus the ved protein pattern does not differ from that of other vent-family proteins. And moreover, its synthesis like that of vox and vent do not depend on pou5f3.
Acknowledgements
We would like to thank Dr. D. Onichtchouk for zebrafish embryos (MZspg793 and WT) kindly donated.
Funding: This study was supported by grant Russian Foundation for Basic Research (14-54 12008) and Russian Federation State contract, registration No. 0104-2018-0022.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Kawahara A, Wilm T, Solnica-Krezel L, et al. Antagonistic role of vega1 and bozozok/dharma homeobox genes in organizer formation. Proc Natl Acad Sci U S A 2000;97:12121-6. [Crossref] [PubMed]
- Kawahara A, Wilm T, Solnica-Krezel L, et al. Functional interaction of vega2 and goosecoid homeobox genes in zebrafish. Genesis 2000;28:58-67. [Crossref] [PubMed]
- Melby AE, Beach C, Mullins M, et al. Patterning the early zebrafish by the opposing actions of bozozok and vox/vent. Dev Biol 2000;224:275-85. [Crossref] [PubMed]
- Imai Y, Gates MA, Melby AE, et al. The homeobox genes vox and vent are redundant repressors of dorsal fates in zebrafish. Development 2001;128:2407-20. [PubMed]
- Ramel MC, Lekven AC. Repression of the vertebrate organizer by Wnt8 is mediated by Vent and Vox. Development 2004;131:3991-4000. [Crossref] [PubMed]
- Belting HG, Wendik B, Lunde K, et al. Pou5f1 contributes to dorsoventral patterning by positive regulation of vox and modulation of fgf8a expression. Dev Biol 2011;356:323-36. [Crossref] [PubMed]
- Shimizu T, Yamanaka Y, Nojima H, et al. A novel repressor-type homeobox gene, ved, is involved in dharma/bozozok-mediated dorsal organizer formation in zebrafish. Mech Dev 2002;118:125-38. [Crossref] [PubMed]
- Zhao J, Lambert G, Meijer AH, et al. The transcription factor Vox represses endoderm development by interacting with Casanova and Pou2. Development 2013;140:1090-9. [Crossref] [PubMed]
- Fabian P, Pantzartzi CN, Kozmikova I, et al. vox homeobox gene: a novel regulator of midbrain-hindbrain boundary development in medaka fish? Dev Genes Evol 2016;226:99-107. [Crossref] [PubMed]
- Kapp L D, Abrams EW, Marlow FL, et al. The integrator complex subunit 6 (Ints6) confines the dorsal organizer in vertebrate embryogenesis. PLoS Genetics 2013;9:e1003822. [Crossref] [PubMed]
- Voronina AS. Translational regulation in early development of eukaryotes. Mol Biol (Mosk) 2002;36:956-69. [PubMed]
- Pshennikova ES, Voronina AS. The proteins of Vent-family and their mRNAs are located in different areas of the tails of Zebrafish and Xenopus embryos. Int J Biochem Cell Biol 2016;79:388-92. [Crossref] [PubMed]
- Pshennikova ES, Tereshina MB, Voronina AS. Expression of vox and vent mRNAs and encoded proteins in zebrafish embryos. Stem Cell Investig 2017;4:60. [Crossref] [PubMed]
- Buitrago-Delgado E, Nordin K, Rao A, et al. Shared Pluripotency Programs Suggest Derivation of Vertebrate Neural Crest from Blastula Cells. Science 2015;348:1332-5. [Crossref] [PubMed]
- Kimmel CB, Ballard WW, Kimmel SR, et al. Stages of embryonic development of the zebrafish. Dev Dyn 1995;203:253-310. [Crossref] [PubMed]
- Voronina A, Pshennikova E. The Vox mRNA and protein expression in zebrafish Pou5f3 MZspg mutant embryos. Stem Cell Investig 2016;3:79. [Crossref] [PubMed]
- Lippok B, Song S, Driever W. Pou5f1 protein expression and posttranslational modification during early zebrafish development. Dev Dyn 2014;243:468-77. [Crossref] [PubMed]
Cite this article as: Pshennikova ES, Voronina AS. The ved protein patterning in zebrafish embryos. Stem Cell Investig 2018;5:17.


