There and back again: amitosis to repopulate a stem cell pool
Proper maintenance of a relatively stable stem cell population is essential for many tissues, within which stem cells and their differentiated progenies function to maintain tissue homeostasis and physiological fitness, especially in response to constantly changing environmental conditions. Failure of such control is tightly linked to tissue degeneration, aging and cancer. Understanding the mechanisms by which the stem cell pool is properly maintained would shed light on the physiological causes of these diseases and facilitate the development of effective therapeutic treatments. In an article published recently in Cell Stem Cell, Lucchetta and Ohlstein investigated such mechanism in the Drosophila intestine, and found that missing stem cells can be replaced by differentiated polyploid cells through a ploidy reduction process known as amitosis (1). This exciting novel study reveals an unexpected mechanism to expand stem cell number in the intestine and respond to more favorable nutritional conditions. It is likely that the underlying principles described in this work will apply to other stem cell populations and other organisms, including in vertebrate tissues.
The adult Drosophila intestinal epithelium is actively maintained by multipotent intestinal stem cells (ISCs), which give rise to secretory enteroendocrine (EE) cells or enteroblast (EB) cells (2-4). While both EEs and young EBs remain diploid, activated EBs undergo endoreplication and cell growth, and ultimately differentiate into large and polyploid (8–16n) absorptive enterocytes (EC). The simplicity of the ISC lineage together with the available cell markers and genetic tools makes the Drosophila intestine an elegant system to study tissue homeostasis in response to nutrient availability, tissue injury and during aging (5-7). A previous study by O’Brien et al. demonstrated that ISCs respond to nutrients during the early developmental growth stage in juvenile flies, by increasing division rate and symmetric cell division and thus increasing the size of the stem cell pool (8). Following these observations, it has been reported that ISC number decreases when adult flies are maintained on a restricted diet and ISC number is recovered when flies are re-fed with protein-rich diet (9) (Figure 1, left). In their recent study, Lucchetta and Ohlstein asked whether the number of ISCs is regulated by drastic reduction in nutritional availability during adulthood. To this end, they developed a complete starvation assay which induces a more rapid and severe loss of ISCs in a sub-region of the posterior midgut. They also found that ISCs are rapidly replaced after flies are re-fed with a protein-rich diet. Through careful quantification of ISC number at different time points following re-feeding, the authors concluded that there is already a significant increase in ISC number by 16 hours of re-feeding. So how are ISCs so rapidly recovered upon re-feeding? Lucchetta and Ohlstein used mitosis quantification and clonal analysis to examine the possibility of a rapid burst of proliferation accompanied by symmetric division, a model similar to the previously described mechanisms involved in juvenile flies. Unexpectedly, they found no significant increases in both mitosis and symmetric cell division 16 hours after re-feeding. It thus seems that increased symmetric division from a few remaining ISCs cannot account for the increase in ISC number during the early recovery stage upon re-feeding.
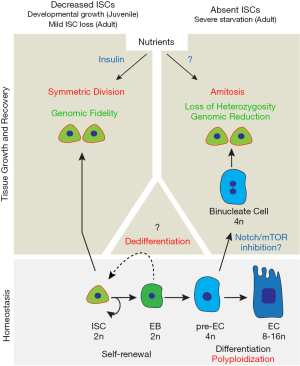
The authors then asked: which cell type(s) could be an alternative source of ISCs? Surprisingly, they found that 4n polyploid cells are the most likely source of new ISCs during the first 16 hours after re-feeding. To examine the cell origin of these new ISCs, they designed two independent experiments. Firstly, they utilized a lineage tracing strategy that labels a subset of EBs and their progenies (EBs, pre-ECs and fully differentiated ECs), but not ISCs and EEs. Unexpectedly, after starvation and re- feeding, a few lineage-labeled ISCs were observed. Importantly, these ISCs are functional and give rise to multi-cell clones, suggesting that new functional ISCs arise from cells in EC lineage, either through diploid EB dedifferentiation or through pre-EC/EC depolyploidization. Secondly, the authors reasoned that, if new functional ISCs are directly generated through ploidy reduction, one can predict that some ISCs will lose heterozygous markers because of random segregation of homologous chromosomes, during the transition from polyploid to diploid cells. They used a combination of fluorescent reporters and genetic mutations to demonstrate that these events do occur during the formation of new ISCs following starvation and re-feeding. All together, these data support the hypothesis that new functional ISCs can be generated through depolyploidization and this study further focuses on the mechanism allowing this process (Figure 1, right). However, the reported experiments do not allow ruling out a significant role for dedifferentiation of diploid EBs in the regeneration of the ISC pool and this alternative mechanism remains to be carefully examined. In addition, the authors found that later during the recovery process, symmetric division of newly created ISCs also plays a homeostatic role to restore a proper stem cell pool.
How could a diploid ISC be generated from a 4n polyploid pre-EC cell? By carefully examining the process in fixed tissue using confocal microscopy, the authors observed key features matching long-existing reports about a process known as amitosis. Amitosis is defined as a specialized cell division in which nuclear invagination separates roughly equal nuclear DNA content into two daughter cells without dissolution of the nuclear membrane, and in the absence of chromatin condensation and mitotic spindle formation. Similar process has been found in several organisms and tissues, including Tetrahymena macronucleus (10), rainbow trout erythrocytes (11), rat trophoblast cells (12) and human fibroblasts (13). However, the mechanism and regulation of this ploidy reduction process remain largely unknown and amitosis has not been described in Drosophila yet. This new study by Lucchetta and Ohlstein opens up the possibility to explore this rather mysterious process in a powerful genetically tractable system, and detailed analyses using a combination of cell markers, advanced imaging and genetic manipulations in the fly gut will certainly provide a better view of the cellular process of amitosis.
Very little is known about the molecular mechanism(s) by which amitosis is regulated. Answers to this question may come from investigating the physiological cues capable of inducing this process in the Drosophila intestine. For example, it would be interesting to examine which dietary component is required under the re-feeding condition. Regarding potential regulatory signaling pathways, the possibilities are almost infinite. Insulin signaling seems to be an attractive first candidate, as local insulin production is required for feeding-induced ISC proliferation and organ growth in newly enclosed flies (8). However, in these juveniles, new ISCs are generated by symmetric division of pre-existing ISCs, suggesting that the mechanisms controlling amitosis in the adult intestine may be completely different. Other pathways are certainly involved in or at least regulated during amitosis. One intriguing observation is that amitosis is found exclusively in the gut region where ISCs are almost depleted by starvation; the idea that the absence of ISCs could serve as an instructive signal for amitosis will be interesting to test. Conversely, it is possible amitosis is actively inhibited in regions of the gut where the number of ISCs is not affected or in conditions where the number of remaining ISCs is sufficient to permit symmetric division to replenish the stem cell pool. Therefore, whether or not a functional ISC is present in the vicinity of a 4n pre-EC cell may modify the patterns used to expand the ISC number in response to feeding between juvenile flies and severely starved adult flies. Obviously, the underlying mechanisms for these proposed regulatory interactions remain completely unknown. Finally, 4n pre-ECs normally go through several rounds of endoreplication and a period of cell growth to increase their ploidy and size; these processes are at least in part controlled by the activity of Notch and TOR signaling (14,15). How are these pathways regulated when amitosis is induced to achieve this seemingly bi-directional fate decision process? It is likely that normal polyploidization and differentiation processes are inhibited to favor amitosis. A better understanding of the mechanism governing polyploidization during the EB-EC differentiation process may shed light on such misregulation and as a consequence the induction of amitosis. We can anticipate that genomic and genetic approaches available in the Drosophila gut will rapidly generate candidates that can be tested to answer all these exciting questions.
What’s the physiological role of amitosis in the intestine? As shown in response to severe ISC loss by starvation, amitosis is induced upon re-feeding to repopulate the ISC pool. Increased amitosis was also observed in response to tissue injury and during aging. It seems that amitosis could benefit the tissue by adapting to stress conditions where most, if not all, ISCs are lost. However, one important but technically challenging question at the moment is how significant role amitosis plays under these conditions. In other words, what happens if amitosis is inhibited? Would this affect sensitivity or regeneration capacity in response to tissue injury or starvation? Also, would impaired amitosis affect aging progress and as a consequence normal lifespan? Such questions would be addressed once genetic manipulations of amitosis become available.
As confirmed by Lucchetta and Ohlstein in their study, amitosis can result in loss of heterozygosity, and as a consequence, generating cells homozygous for deleterious mutations. Considering the tight association between polyploidy, aneuploidy and tumorigenesis (16,17), as the authors point out, such inability to reproducibly pass on genomic information during amitotic division may initiate and facilitate malignant transformation.
It is obvious from this short discussion of the study by Lucchetta and Ohlstein that, like all great scientific advances, it produces many more questions than it provides answers. Regardless of all the open questions, they describe for the first time that amitosis could act as an alternative mechanism to replace somatic stem cells in the Drosophila intestine, adding yet another layer of cellular plasticity in this tissue. It provides a unique system to investigate the physiological role and the underlying mechanism and regulation of amitosis. Such mechanisms are likely to be conserved in other stem cell populations and organisms, and be involved in tumorigenesis and the aging process. Once again Drosophila finds itself at the forefront of innovative research in cell biology.
Acknowledgements
We would like to thank all members in the Biteau laboratory for helpful discussion and critical reading of the manuscript.
Funding: Work in the Biteau laboratory is supported by the National Institute of General Medical Sciences (R01GM108712) and the Ellison Medical Foundation (AG-NS-0990-13).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Lucchetta EM, Ohlstein B. Amitosis of Polyploid Cells Regenerates Functional Stem Cells in the Drosophila Intestine. Cell Stem Cell 2017;20:609-20.e6. [Crossref] [PubMed]
- Micchelli CA, Perrimon N. Evidence that stem cells reside in the adult Drosophila midgut epithelium. Nature 2006;439:475-9. [Crossref] [PubMed]
- Ohlstein B, Spradling A. The adult Drosophila posterior midgut is maintained by pluripotent stem cells. Nature 2006;439:470-4. [Crossref] [PubMed]
- Biteau B, Jasper H. Slit/Robo signaling regulates cell fate decisions in the intestinal stem cell lineage of Drosophila. Cell Rep 2014;7:1867-75. [Crossref] [PubMed]
- Biteau B, Hochmuth CE, Jasper H. Maintaining tissue homeostasis: dynamic control of somatic stem cell activity. Cell Stem Cell 2011;9:402-11. [Crossref] [PubMed]
- Gervais L, Bardin AJ. Tissue homeostasis and aging: new insight from the fly intestine. Curr Opin Cell Biol 2017;48:97-105. [Crossref] [PubMed]
- Li H, Jasper H. Gastrointestinal stem cells in health and disease: from flies to humans. Dis Model Mech 2016;9:487-99. [Crossref] [PubMed]
- O'Brien LE, Soliman SS, Li X, et al. Altered modes of stem cell division drive adaptive intestinal growth. Cell 2011;147:603-14. [Crossref] [PubMed]
- McLeod CJ, Wang L, Wong C, et al. Stem cell dynamics in response to nutrient availability. Curr Biol 2010;20:2100-5. [Crossref] [PubMed]
- Endo M, Sugai T. Amitotic division of the macronucleus in Tetrahymena thermophila: DNA distribution by genomic unit. Zoolog Sci 2011;28:482-90. [Crossref] [PubMed]
- Wang B, Liu Y, Chen X, et al. Amitosis-like nuclear division in erythrocytes of triploid rainbow trout Oncorhynchus mykiss. J Fish Biol 2010;76:1205-11. [Crossref] [PubMed]
- Zybina EV, Zybina TG. Modifications of nuclear envelope during differentiation and depolyploidization of rat trophoblast cells. Micron 2008;39:593-606. [Crossref] [PubMed]
- Walen KH. Budded karyoplasts from multinucleated fibroblast cells contain centrosomes and change their morphology to mitotic cells. Cell Biol Int 2005;29:1057-65. [Crossref] [PubMed]
- Kapuria S, Karpac J, Biteau B, et al. Notch-mediated suppression of TSC2 expression regulates cell differentiation in the Drosophila intestinal stem cell lineage. PLoS Genet 2012;8:e1003045. [Crossref] [PubMed]
- Perdigoto CN, Schweisguth F, Bardin AJ. Distinct levels of Notch activity for commitment and terminal differentiation of stem cells in the adult fly intestine. Development 2011;138:4585-95. [Crossref] [PubMed]
- Davoli T, de Lange T. The causes and consequences of polyploidy in normal development and cancer. Annu Rev Cell Dev Biol 2011;27:585-610. [Crossref] [PubMed]
- Duncan AW. Aneuploidy, polyploidy and ploidy reversal in the liver. Semin Cell Dev Biol 2013;24:347-56. [Crossref] [PubMed]
Cite this article as: Meng FW, Biteau B. There and back again: amitosis to repopulate a stem cell pool. Stem Cell Investig 2017;4:82.


