Epigenetics meets metabolism through PHB-mediated histone H3.3 deposition by HIRA
The regulation of chromatin, the histone-mediated packaged form of DNA in the eukaryotic nucleus, underlies the epigenetic control of gene expression (1). Epigenetic regulators include DNA and histone modifying enzymes, proteins that specifically target native or modified DNA and histones, non-coding RNA molecules and histone chaperone and ATP-dependent remodeling complexes that deposit, move or evict canonical or variant histones. While the mode of chromatin targeting of many epigenetic factors has been delineated (2), the mechanism of histone chaperone complex recruitment for specific histone deposition is not understood. One such histone chaperone complex contains the proteins HIRA, Ubinuclein-1 (UBN1), CABIN1 and transiently associated ASF1 to deposit the histone H3.3 variant at the promoters of active and poised genes, bodies of active genes, developmental genes, active enhancers and sites of DNA and chromatin damage and repair (3) (Figure 1). A recent study demonstrates that the single stranded DNA-binding protein replication protein A (RPA) cooperates with the HIRA complex to deposit newly synthesized H3.3 during the G1 phase of the cell cycle at gene transcription regulatory elements (4) (Figure 1). However, how HIRA mediates H3.3 deposition independent of the cell cycle remained unknown. A recent report by Zhu et al. sheds some light on this area (5).
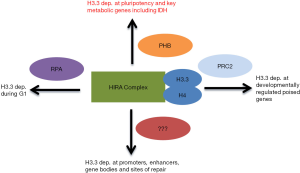
Zhu et al. carried out a genome-wide siRNA screen for pluripotency/self-renewal factors in human embryonic stem cells (hESCs), leading to the identification of prohibitin (PHB) as one such factor (Figure 1). PHB is an evolutionarily conserved and ubiquitous expressed protein that has previously been implicated in mitochondrial function and morphology, cell signaling and transcriptional regulation (6). To validate the importance of PHB as a pluripotency factor in hESCs, Zhu et al. observed that PHB expression was high in undifferentiated hESCs and showed that PHB knock down (KD) induced extensive differentiation in hESCs. A global gene expression analysis of a PHB KD in hESCs revealed that upregulated genes are associated with cell differentiation and organismal development, while down-regulated genes are associated with gene ontology terms chromatin modification, glucose metabolism and RNA processing, together suggesting that PHB might mediate self-renewal through maintaining the hESC chromatin landscape and metabolic state. The connection of PHB to chromatin regulation was reinforced by the finding that, compared to control cells, PHB KD hESCs had higher levels of mono-, di-, and tri-methylated histone 3 lysine 4 (H3K4), histone 3 lysine 27 (H3K27) as well as tri-methylated histone 3 lysine 9 (H3K9) and histone 3 lysine 36 (H3K36).
To understand more about the molecular function of PHB, Zhu et al. searched for associated proteins through immunoprecipitation and mass spectrometry, leading to the identification of HIRA as an associated factor. This interaction was validated through co-immunoprecipitation (co-IP) experiments and subsequently mapped, through deletion analysis to primarily an N-terminal WD40 repeat of HIRA and a central conserved PHB domain of PHB. Interaction of these two proteins was subsequently demonstrated with recombinant proteins, thus implicating a direct interaction.
Given that HIRA forms a complex with UBN1 and CABIN1, the authors asked if PHB could interact with these other subunits of the HIRA complex. As expected, the authors found that PHB could co-IP each of the HIRA complex subunits from HEK293T cells. Moreover, gel filtration analysis indicated that PHB and each of the HIRA complex subunits and H3.3 coexist in the same gel filtration fraction. PHB was also found in HIRA free fractions, suggesting that a proportion of PHB exists unbound to HIRA. To validate the functional importance of the PHB/HIRA complex interaction in expression of pluripotency genes in hESC, Zhu et al. demonstrated that, like PHB KD, HIRA−/− hESCs displayed a differentiated cell morphology and had reduced expression levels of core pluripotency factors, with concomitant upregulation of various lineage marker genes, and exhibited the same perturbed histone modification patterns as the PHB KD.
Metabolic states of the cell have been shown to impact the role of epigenetic mechanisms in determining stem cell fate (7). One example of this is the metabolite α-ketoglutarate (α-KG), a TCA cycle intermediate generated by isocitrate dehydrogenases (IDHs) (8). α-KG is also an essential cofactor of dioxygenases, such as jumonji (JMJ) family histone demethylases (9) and ten-eleven translocation (TET) family methylated DNA hydroxylases (10). While the TET enzymes can mediate one or more steps in a multi-step DNA demethylation process, at least some of the JMJ enzymes are bona fide histone demethylases.
Given that the elevated histone methylation phenotype induced by PHB or HIRA KD in hESCs resembled the histone methylation alterations observed with IDH1 mutations and reduced α-KG production, Zhu et al. asked whether disruption of the PHB and HIRA complexes could affect the IDH/α-KG metabolic pathway. They found that PHB or HIRA KD in hESCs decreased IDH and α-KG levels, and that the levels of α-KG correlated inversely with the same methylated histones after KD of PHB and HIRA levels in hESCs. Taken together, this data links PHB and the HIRA histone chaperone complex to the expression of IDH genes and hence production of α-KG and histone demethylation (Figure 1).
To understand how PHB and HIRA are functionally connected to IDH gene expression and thus α-KG levels, Zhu et al. carried out genome wide H3.3 ChIP-seq in hESCs stably expressing H3.3B-HA treated with NT (control), PHB, or HIRA siRNA. As anticipated, they found that H3.3 is selectively enriched at transcriptionally active genes over less active genes. In addition, KD of PHB or HIRA reduced H3.3 levels at active genes. Bioinformatics analysis of the H3.3 regions with >1.5 reduction of ChIP-seq reads in both PHB and HIRA KD cells indicated that these regions were enriched at genes involved in cellular metabolic processes, the cell cycle, chromatin binding, and transcription regulation. Subsequent ChIP-seq of PHB and HIRA demonstrated that these proteins were simultaneously recruited to IDH genes, as well as other pluripotency and development-associated genes in hESCs, such as OCT4 and NANOG (11). Moreover, KD of PHB and HIRA decreased H3.3 deposition at all these genes, and decreased the active transcription markers H3K4me3, RNA polymerase II S5P and elongation RNA polymerase II S2P. This data links PHB to HIRA-mediated H3.3 deposition to activation of transcription of IDH genes to produce α-KG, a cofactor for histone demethylation.
To further investigate the temporal relationship between PHB function, the epigenetic-metabolic circuitry and hESC identify, Zhu et al. characterized PHB-depleted hESC cells from 0 to 4 days. These studies suggested that a reduction of HIRA complex components could be the first event that triggers loss of hESC identity. This finding is consistent with earlier studies by the authors demonstrating that PHB increases the cellular stability of HIRA complex components.
Together, the studies by Zhu et al. demonstrate that PHB is an essential regulator of self-renewal in hESCs and works, at least in part, through interaction with the HIRA histone chaperone complex to deposit the H3.3 histone variant to facilitate expression of IDH genes to produce α-KG, an important metabolite for normal chromatin landscapes and hESC identity. This is an important finding in the field of hESC pluripotency factors as it identifies PHB as a new player in this arena, and points to its cooperation with the HIRA histone chaperone complex in mediating this effect. However, the observations that PHB is also localized to the mitochondria and that α-KG supplementation could rescue some but not all of the changes caused by PHB deficiency, argues for other roles of PHB, as well α-KG, which awaits further studies for resolution.
The studies reported here also implicate an important new role for the HIRA histone chaperone complex in maintaining self-renewal in hESCs. The HIRA histone chaperone complex has previously been described to deposit histone H3.3 at active and poised promoters, active gene bodies, developmental genes and sites of DNA and chromatin damage and repair, and to play an important role in control of chromatin dynamics in non-proliferating senescent cells (12). The important role of HIRA in contributing to the maintenance of both senescence and hESCs through histone H3.3 deposition implicates a broader role of the HIRA histone chaperone complex and the histone H3.3 histone variant in maintaining cell identity. Consistent with this, Allis and coworkers previously revealed a role for HIRA in lineage determination in ESCs (13).
The mechanism for how PHB contributes to HIRA-mediated histone H3.3 deposition is not clear. The studies by Zhu et al. demonstrate that HIRA and PHB directly interact. Given that PHB has been shown to interact with DNA-binding transcription factors such as nuclear receptors (14,15), it is possible that it might mediate the recruitment of the HIRA histone chaperone complex to specific chromatin regions for histone H3.3 deposition. Gel filtration analysis of proteins from hESC cells reveals a fraction that contains PHB and each of the components of the HIRA complex and histone H3.3. However, the elution pattern of each of the proteins does not precisely co-migrate, suggesting that both HIRA and PHB are also part of other complexes. Strikingly, PHB does co-migrate exactly with the CABIN1 subunit of the HIRA complex, suggesting that PHB may form an obligate complex with CABIN1, a subunit of the HIRA complex that has been implicated to bind several transcription factors including MEF2B, MEF2D and SIN3A (16). Intriguingly, the role of CABIN1 in H3.3 deposition is also poorly defined. Perhaps a concerted mechanistic analysis of both proteins and their role in the HIRA complex will jointly shed light on their role in H3.3 deposition.
As with many such studies, the findings of Zhu et al. generate several unresolved questions. First, as shown previously in other cell types, the distribution of H3.3 across the genome in hESCs is broad, present at many active promoters, gene bodies and developmentally poised promoters (13,17-21). Yet, the phenotype caused by HIRA and PHB inactivation is largely abrogated by the cell permeable α-KG mimetic, octyl-α-KG. This supports the authors’ interpretation that most of the effects of HIRA and PHB inactivation are due to downregulation of IDH enzymes. The reasons for the specific funneling of the consequences of H3.3 depletion via a single metabolic axis are curious and important to address. Second, the authors note the differences between their results in hESCs and previous reported results in mouse ESCs (mESCs) (13,17). In mESCs, inactivation of histone H3.3 has only a modest phenotype and affects only a very small number of genes (13,17). The effects reported here in hESCs are much greater. The modest effects previously reported in proliferating mESCs were previously suggested to be due to functional redundancy of HIRA-mediated nucleosome assembly with other DNA replication coupled pathways (17). The marked effects reported here in, presumably proliferating, hESCs do not support this view. Instead, the authors suggest that the difference is due to the more primitive nature of mESCs, compared to hESCs. Resolution of this difference will shed important light on comparative nucleosome and chromatin regulatory pathways in hESCs and mESCs. Third, mutation of IDH enzymes, dysregulation of α-KG and epigenetic control is implicated in some human cancers, including AML and glioblastoma (22). The key role for HIRA and PHB in production of α-KG tempts speculation that this chaperone complex, including UBN1 and CABIN1, might also be involved in cancer etiology via the same pathway. Although dysregulation of PHB has been linked to cancer (23), according to cBioPortal mostly by gene amplifications, and histone H3.3 is a target of specific mutations in some cancers, e.g., pediatric gliomas (24), there is no obvious connection between these mutations and the IDH/α-KG pathway reported here. Conceivably, H3.3 mutations that prevent histone methylation, such as H3.3K27M, might perturb the global balance of histone methylation in a way that mimics HIRA and PHB inactivation. For example, H3.3K27M mutations paradoxically increase H3K27me3 at some tumor suppressor loci (25), as might HIRA and/or PHB inactivation. Previous studies in a mouse model have implicated HIRA in tumor suppression (12).
A particularly interesting aspect of the studies of Zhu et al. is the connection that is made between the epigenetic factor HIRA and the production of the metabolite α-KG, a cofactor that in turn is used by JmjC-domain containing histone demethylases to mediate epigenetic regulation. Other examples of the regulation of epigenetic status by metabolites include acetyl-CoA as a substrate of histone acetyltransferases, SAM as a substrate of lysine and arginine histone methyltransferases and DNA methyl transferases and NAD+ as a substrate of the sirtuin family of histone deacetylases (26). It appears that the epigenetic-metabolic circuit mediated by HIRA and PHB in human ESCs may only be yet another connection that links epigenetics, metabolism and cell identity. Finally, via on-going DNA replication-independent deposition in non-proliferating cells, histone H3.3 accumulates with age in post-mitotic tissues (27,28), and is required for longevity phenotypes in worms (29). It will be interesting to determine whether this, in turn, affects the epigenetic-metabolic axis revealed here, leading to age-dependent epigenetic-metabolic disruption.
Acknowledgements
Funding: This work was supported by the National Institutes of Health grant (No: P01 AG031862) to R Marmorstein, PD Adams.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell 2007;128:635-8. [Crossref] [PubMed]
- Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet 2016;17:487-500. [Crossref] [PubMed]
- Ricketts MD, Marmorstein R. A Molecular Prospective for HIRA Complex Assembly and H3.3-Specific Histone Chaperone Function. J Mol Biol 2016. [Epub ahead of print]. [Crossref] [PubMed]
- Zhang H, Gan H, Wang Z, et al. RPA Interacts with HIRA and Regulates H3.3 Deposition at Gene Regulatory Elements in Mammalian Cells. Mol Cell 2017;65:272-84. [Crossref] [PubMed]
- Zhu Z, Li C, Zeng Y, et al. PHB Associates with the HIRA Complex to Control an Epigenetic-Metabolic Circuit in Human ESCs. Cell Stem Cell 2017;20:274-89. e7.
- Mishra S, Murphy LC, Murphy LJ. The Prohibitins: emerging roles in diverse functions. J Cell Mol Med 2006;10:353-63. [Crossref] [PubMed]
- Ryall JG, Cliff T, Dalton S, et al. Metabolic Reprogramming of Stem Cell Epigenetics. Cell Stem Cell 2015;17:651-62. [Crossref] [PubMed]
- Su X, Wellen KE, Rabinowitz JD. Metabolic control of methylation and acetylation. Curr Opin Chem Biol 2016;30:52-60. [Crossref] [PubMed]
- Franci G, Ciotta A, Altucci L. The Jumonji family: past, present and future of histone demethylases in cancer. Biomol Concepts 2014;5:209-24. [Crossref] [PubMed]
- Kohli RM, Zhang Y. TET enzymes, TDG and the dynamics of DNA demethylation. Nature 2013;502:472-9. [Crossref] [PubMed]
- Kulcenty K, Wroblewska J, Mazurek S, et al. Molecular mechanisms of induced pluripotency. Contemp Oncol (Pozn) 2015;19:A22-9. [Crossref] [PubMed]
- Rai TS, Cole JJ, Nelson DM, et al. HIRA orchestrates a dynamic chromatin landscape in senescence and is required for suppression of neoplasia. Genes Dev 2014;28:2712-25. [Crossref] [PubMed]
- Banaszynski LA, Wen D, Dewell S, et al. Hira-dependent histone H3.3 deposition facilitates PRC2 recruitment at developmental loci in ES cells. Cell 2013;155:107-20. [Crossref] [PubMed]
- Kurtev V, Margueron R, Kroboth K, et al. Transcriptional regulation by the repressor of estrogen receptor activity via recruitment of histone deacetylases. J Biol Chem 2004;279:24834-43. [Crossref] [PubMed]
- Murphy LC, Leygue E, Niu Y, et al. Relationship of coregulator and oestrogen receptor isoform expression to de novo tamoxifen resistance in human breast cancer. Br J Cancer 2002;87:1411-6. [Crossref] [PubMed]
- Youn HD, Liu JO. Cabin1 represses MEF2-dependent Nur77 expression and T cell apoptosis by controlling association of histone deacetylases and acetylases with MEF2. Immunity 2000;13:85-94. [Crossref] [PubMed]
- Goldberg AD, Banaszynski LA, Noh KM, et al. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 2010;140:678-91. [Crossref] [PubMed]
- Mito Y, Henikoff JG, Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat Genet 2005;37:1090-7. [Crossref] [PubMed]
- Pchelintsev NA, McBryan T, Rai TS, et al. Placing the HIRA histone chaperone complex in the chromatin landscape. Cell Rep 2013;3:1012-9. [Crossref] [PubMed]
- Jin C, Felsenfeld G. Distribution of histone H3.3 in hematopoietic cell lineages. Proc Natl Acad Sci U S A 2006;103:574-9. [Crossref] [PubMed]
- Ahmad K, Henikoff S. The histone variant h3.3 marks active chromatin by replication-independent nucleosome assembly. Mol Cell 2002;9:1191-200. [Crossref] [PubMed]
- Dang L, Yen K, Attar EC. IDH mutations in cancer and progress toward development of targeted therapeutics. Ann Oncol 2016;27:599-608. [Crossref] [PubMed]
- Koushyar S, Jiang WG, Dart DA. Unveiling the potential of prohibitin in cancer. Cancer Lett 2015;369:316-22. [Crossref] [PubMed]
- Yuen BT, Knoepfler PS. Histone H3.3 mutations: a variant path to cancer. Cancer Cell 2013;24:567-74. [Crossref] [PubMed]
- Chan KM, Fang D, Gan H, et al. The histone H3.3K27M mutation in pediatric glioma reprograms H3K27 methylation and gene expression. Genes Dev 2013;27:985-90. [Crossref] [PubMed]
- Etchegaray JP, Mostoslavsky R. Interplay between Metabolism and Epigenetics: A Nuclear Adaptation to Environmental Changes. Mol Cell 2016;62:695-711. [Crossref] [PubMed]
- Maze I, Wenderski W, Noh KM, et al. Critical Role of Histone Turnover in Neuronal Transcription and Plasticity. Neuron 2015;87:77-94. [Crossref] [PubMed]
- Pina B, Suau P. Changes in histones H2A and H3 variant composition in differentiating and mature rat brain cortical neurons. Dev Biol 1987;123:51-8. [Crossref] [PubMed]
- Piazzesi A, Papic D, Bertan F, et al. Replication-Independent Histone Variant H3.3 Controls Animal Lifespan through the Regulation of Pro-longevity Transcriptional Programs. Cell Rep 2016;17:987-96. [Crossref] [PubMed]
Cite this article as: Marmorstein R, Adams PD. Epigenetics meets metabolism through PHB-mediated histone H3.3 deposition by HIRA. Stem Cell Investig 2017;4:46.


