Will targeting PI3K/Akt/mTOR signaling work in hematopoietic malignancies?
Introduction
The components of the phosphatidylinositol 3-kinase/protein kinase B/mammalian target of rapamycin (PI3K/Akt/mTOR) pathway are frequently mutated in human cancers. This pathway controls the cell cycle, survival, metabolism, and genomic instability of cells that are also the main hallmarks of cancer abnormality (1). Activated PI3K/Akt/mTOR signaling is frequently elicited by the direct mutational activation along the pathway, amplification of genes encoding key components such as PIK3CA (2-8), AKT1 (9) or the loss of tumor suppressor such as PTEN (10-13). Many drugs targeting this pathway, alone or in combination with other existing or developing drugs, are currently in clinical trials both for solid tumors and hematologic neoplasms with mediocre efficacies. Here in this mini-review, we will examine the pathway components and their functions, assess the available clinical and experimental data, and discuss the possible direction we may undergo in developing targeting drugs.
Components of the PI3K/Akt/mTOR signaling pathway and cancer
PI3Ks and PTEN
In mammals, the PI3K family consists of three lipid kinase classes: class I, II and III. Class I PI3Ks is further divided into two subclasses: subclass IA (PIK3CA, PIK3CB and PIK3CG) and subclasses IB (PIK3CD). Class I PI3Ks are heterodimers that consist of a catalytic and a regulatory subunit. There are four catalytic isoforms (PIK3CA, PIK3CB, PIK3CG and PIK3CD) and several regulatory subunits (PIK3R1, PIK3R2, PIK3R3, PIK3R5 and PIK3R6) in class I PI3Ks. Among the four catalytic isoforms of class I PI3K, only PIK3CA is frequently mutated in more than 30% of various human solid tumors (14). Mutations of PIK3CB, the ubiquitously expressed gene, are rarely seen (15). Class I PI3K regulatory subunits such as PIK3R1 and PIK3R2, were found in cancer cells and resulted in PI3K activation (16,17). Class II PI3Ks are not well defined at present (18). Class III PI3K refers to vacuolar protein sorting 34 (VPS34: PIK3C3), which is a critical regulator of autophagy (19). The phosphatase and tensin homolog deleted on chromosome 10 (PTEN), is the first phosphatase identified as a tumor suppressor. PTEN is frequently silenced or mutated in almost all cancers (20).
PDK1 (PDKP1) and Akt
3-phosphoinositide-dependent protein kinase 1 (PDK1 or PDKP1) and protein kinase B (Akt), which belong to serine/threonine (Ser/Thr) kinase, are both members of the AGC kinase family (cAMP-dependent, cGMP-dependent and protein kinase C) (21,22). PDK1 overexpression was observed in breast carcinoma (23), prostate cancer (24), esophageal squamous cell carcinoma (25), melanoma (26), and AML (27), and correlated with tumor invasion and poor prognosis (28). The three main isoforms of Akt are termed Akt1, Akt2, and Akt3. (22). Akt1 was overexpressed in primary human gastric adenocarcinoma (29), Akt2 was overexpressed in ovarian, gastric, breast and pancreatic cancer (30-32). Akt3 was overexpressed in primary melanomas, ovarian tumors, and prostate cancers (33-35). Somatic mutations in Akt1 occurred in a small number of human breast, ovarian, and colorectal cancers (36). Increased copy number of chromosomal region 1q44 (where AKT3 is located) was observed in hepatocellular carcinomas and glioblastomas (37,38).
FoxOs
The FoxO transcription factors are considered as tumor suppressors that belong to the forkhead box family of transcription regulators (39). The FoxO family contains four members: FoxO1, FoxO3, FoxO4 and FoxO6 (40). The FoxOs are involved in multiple signaling pathways and play critical roles in a variety of biological processes including cell-cycle arrest, differentiation, apoptosis, metabolism and stress resistance (41).
TSC complex and Rheb
The TSC complex, comprising of TSC1, TSC2 and TBC1D7, is GTPase-activating proteins (GAPs) of Ras homologue enriched in brain (Rheb) (42-44). Tuberous sclerosis (TSC), which is caused by mutation of TSC1 (encoding hamartin) or TSC2 (encoding tuberin) genes, is a rare genetic disorder characterized by tumor formation in multiple organs (45). Two Rheb family members, Rheb1and Rheb2 (also known as RhebL1) have been identified in mammals. Rheb1 was overexpressed in various cancers, such as non-small cell lung cancer, liver cancer, bladder cancer, breast cancer, head and neck cancers, prostate cancer, and acute myeloid leukemia (AML) (46-52). The human cancer genome data analysis confirmed existence of recurrent Rheb1 mutations (53). Rheb1 deletion augmented the apoptosis of AML cells, and rapamycin combined with Rheb1 deletion further increased AML cell apoptosis in a MLL-AF9 induced mouse model (51).
mTOR
mTOR is an evolutionarily conserved Ser/Thr kinase that senses and responds to various signals to regulate cell growth, cell survival and other multiple biological processes in eukaryotes (54). It belongs to PI3K-related kinases (PI3KKs) (55). mTOR interacts with multi-proteins to form two distinct complexes, designated mTOR complex 1 (mTORC1) and mTOR complex 2 (mTORC2). Raptor and PRAS40 belong specifically to mTORC1, while Rictor, mSin1 and Protor are specific components of mTORC2 (56,57). mTOR complexes play critical roles in both normal hematopoiesis and leukemogenesis (58,59). mTOR hyper-activation was observed in various cancers of 123 individual studies in the cBioportal (http://www.cbioportal.org/, data not shown).
Interactions of these components
Transmembrane tyrosine kinase growth factor receptors (TKGFR) such as insulin-like growth factor 1 receptor (IGF1R), ErbB family receptors and fibroblast growth factor receptors (FGFRs), and G-protein-coupled receptors (GPCR) can initiate the PI3K/Akt/mTOR signaling pathway upon receiving stimulation cues. The activated PI3K is then translocated to the plasma membrane, resulting in the phosphorylation of PIP2 to PIP3. PTEN, a negative regulator of this process, dephosphorylates PIP3 to PIP2. Subsequently, PIP3 activates PDK1 and Akt. The activated PDK1 then phosphorylates Akt at T308 and the activated mTORC2 phosphorylated Akt at S473. The activated Akt phosphorylates TSC2, which inhibits TSC1-TSC2 complex. As a GAP for Rheb, TSC1-TSC2 complex can convert Rheb from its active GTP-binding form to inactive GDP-binding form through GTP hydrolysis. Following the inhibition of TSC1-TSC2 complex, Rheb activates mTORC1, leading to protein synthesis and cell growth via 4E-BP1 and S6K1 (Figure 1). The phosphorylated S6K1 inhibits the insulin receptor substrate-1 (IRS-1), which promotes IRS-1 degradation and reduces the PI3K signaling to finish the feedback loop (60) (Figure 1).
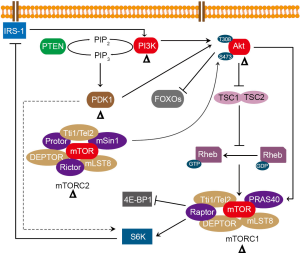
Inhibitors targeting PI3K/Akt/mTOR signaling pathway for hematologic malignancies
The PI3K/Akt/mTOR signaling pathway has been demonstrated to be frequently dysregulated in hematologic malignancies and associated with poor outcome. The constitutive activation of PI3K/Akt/mTOR signaling was observed in 50-80% of AML cases (61-63) and in 87.5% of T cell-acute lymphoblastic leukemia (T-ALL) cases (64). The pathway was also activated in chronic myelocytic leukemia (CML) (65), chronic lymphocytic leukemia (CLL) (66), multiple myeloma (MM) (67,68), and high-risk myelodysplastic syndrome (MDS) (69). Therefore, much effort has been devoted to developing precision medicine to this pathway for treating hematological malignancies. There are currently four main classes of inhibitors that target the PI3K/Akt/mTOR signaling pathway: PI3K inhibitors, Akt inhibitors, mTOR inhibitors and dual PI3K-mTOR inhibitors (70). PI3K inhibitors consist of pan-PI3K inhibitors and isoform-selective inhibitors. The former are capable of inhibiting all four isoforms of Class I PI3K, while the latter specifically target only one isoform of PI3K (71). Next-generation inhibitor that could overcome mTOR resistance mutations has also been developed (72).
Fransecky et al. reviewed the preclinical and clinical trials of these aforementioned inhibitors in AML, B-ALL, B-CLL, T-ALL, non-Hodgkin lymphoma (NHL), and MDS, but found that the clinical trial data are rather disappointing (73). In addition to the four main inhibitors that targeting the PI3K/Akt/mTOR signaling pathway, many other inhibitors are also in development. Studies showed that PDK1 inhibitors could inhibit human AML cell growth in vitro (27,74,75). This is supported by experimental evidence that PDK1 deletion in mice could prolong the survival of MLL-AF9 induced AML mice when compared with the control (76). Interestingly, the dual PI3K/PDK1 inhibitors were more cytotoxic to T-ALL cell lines and primary patients leukemia cell samples when compared with the pan-PI3K inhibitor, Akt inhibitor, mTORC1 inhibitors, and mTORC1/mTORC2 inhibitors used in the same study (77). While so many inhibitors targeting this pathway have been tested, the clinical trial results are rather mediocre in contrast to the convincing preclinical data with the same drugs.
Genetic alterations of PI3K/Akt/mTOR signaling pathway in hematologic malignancies
Why the efficacy of inhibitors targeting this pathway was not as good as expected in contrast to the initial experimental anti-malignancy results? There are three possible interpretations. First, molecules in this pathway have crosstalk(s) and/or feedback loop(s). For example, suppression of mTORC1 inhibits downstream S6K1, inducing IRS-1-dependent negative feedback that promotes activation of PI3K/Akt (60,78). Second, inhibitors developed currently could not totally abrogate the full function of a particular targeted molecule, and the downstream molecules are not always completely inhibited. For example, phosphorylation of 4E-BP1 was not much affected by rapalogs or mTORC1 inhibitors in AMLs (79). Furthermore, cellular metabolic status also affect the inhibition result as Medvetz and colleagues demonstrated that mTORC1 hyper-activation generates metabolic vulnerabilities that can be therapeutically targeted while mTORC1 inhibition alleviates these metabolic vulnerabilities (80). Third, drug-resistance mutations may occur quickly upon mTOR inhibitor usage in breast cancer cell line (72). Given these, it is essential to further explore the molecular signatures of PI3K/Akt/mTOR in hematological malignancies in order to better the design of inhibitors.
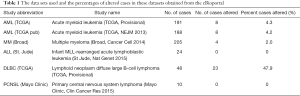
Previous studies showed that the constitutive activation of the PI3K/Akt/mTOR signaling pathway was due to existing mutations of FLT3 in AML (63), BCR-ABL fusion protein in CML (65), and K-RAS/N-RAS mutations or PTEN mutations in MM (67,68). To further explore the dysregulations of this pathway in hematologic neoplasms, we analyzed the mutation types and copy number alterations (CNA) of PI3K/Akt/mTOR signaling pathway genes in hematologic malignancies using the cBioportal for Cancer Genomics (http://www.cbioportal.org/), which provides large-scale cancer genomics data sets. The Cancer Genome Atlas (TCGA) which includes data of AML (from TCGA Provisional, 200 cases and TCGA, NEJM, 2013, 200 cases), Infant MLL-rearranged acute lymphoblastic leukemia (St Jude, Nat Genet 2015, 24 cases), MM (Broad, Cancer Cell 2014, 205 cases), lymphoid neoplasm diffuse large B-cell lymphoma (DLBC, TCGA, Provisional, 48 cases), and primary central nervous system lymphoma (PCNSL, Mayo Clinic, Clin Cancer Res, 2015, 10 cases), were used for mutations analysis (81). We found that the percentages of genetically altered cases in the AML (TCGA), AML (TCGA, pub), MM (broad) and DLBC (TCGA) were 4.3%, 4.2%, 2% and 47.9% respectively, while those of ALL (St. Jude) and PCNSL (Mayo Clinic) were both 0% (Table 1).
Full table
We then analyzed the mutation types of the 17 selected genes in the PI3K/Akt/mTOR signaling pathway in these cases with mutations. In AML (TCGA), there are 1% PIK3R2 amplification, 1% PTEN deep deletion, 1% MTOR fusion and missense mutation and 3% RHEB deep deletion. In AML (TCGA, pub), there are 1% PIK3R2 amplification, 1% PTEN deep deletion, 1% MTOR deep deletion and 3% RHEB deep deletion. While there are missense mutations of AKT1, MTOR, TSC1 and MLST8 in MM (Broad), PTEN deletions and hotspot mutations of PIK3CA and AKT1 are absent in this MM dataset (82,83). Surprisingly, more mutations are seen in DLBC (TCGA) that involved at least 14 genes in the PI3K/Akt/mTOR signaling pathway (Figure 2). Among these detected hematological malignancies, PIK3R2 amplification and PTEN deep deletion in AML, as well as PIK3CA, PDPK1, MLST8 and RICTOR amplification in DBLC could contribute to the activation of this pathway. However, they only constitute a small subset of hematological malignancies except for DLBC dataset.

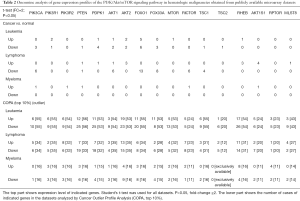
Subsequently, we compared the expression of the 17 selected genes in the PI3K/Akt/mTOR signaling pathway in the hematological malignancies VS normal counterparts existed in Oncomine (www.oncomine.org), which is a cancer microarray database that provides gene expression profiles. We found that the expression of PIK3R1, MTOR and RHEB in leukemia, that of PDPK1, AKT2, MTOR2, RHEB, AKT1S1 and MLST8 in lymphoma, and that of PI3CA, PIK3R2, RICTOR and AKT1S1 in myeloma were all increased when compared with their normal controls (P<0.05, fold-change ≥2), consistent with the activation of the PI3K/Akt/mTOR signaling pathway in hematological diseases. Similarly, Cancer outlier profile analysis (COPA) showed increased MLST8 expression in a small subset of lymphoma (4/27) and enhanced RICTOR expression in a small subset of myeloma (2/11) (P<0.05, fold-change ≥2) (Table 2). Therefore, inhibitors targeting PI3K/mTOR/Akt signaling pathway could inhibit cancer cell growth in a very small proportion of patients with hematological malignancies.
Full table
Conclusions and perspectives
Mutations in genes from PI3K/mTOR/Akt pathway resulting in the activation of this pathway comprise only a very small subset of hematological malignancies. Inhibitors to the specific gene products in the pathway failed to show high efficacy in all hematological malignancies thus far. This is probably caused by the remaining activity of the PI3K/Akt/mTOR pathway owing to partial efficacy of the inhibitors, the upregulation of regulatory feedback loop, or the adaptive mutation in response to inhibitors. In view of this, the search for new type of inhibitors that are more effective, precisely targeted and less toxic in the PI3K/Akt/mTOR signaling pathway is urgently needed. More importantly, gene products that regulate the up- or down-stream of the PI3K/Akt/mTOR signaling pathway may provide an alternative for consideration when designing drugs targeting hematological malignancies. In this case, aiming AMPK and MAPK pathways could be an attractive approach. In addition, gene expression is also regulated by additional factors such as epigenetic elements, microRNAs or lncRNAs, thus searching epigenetic changes that regulate these pathways in hematological malignancies may also expand our horizon for curing diseases.
Although the mutation type and expression level of genes in PI3K/AKT/mTOR signaling were widely accessible online, the profiles for their protein expression level data were not available due to probably the lack of such data. Since the protein level is considered to be the most valuable data to corroborate its gene expression level and provide more valid information for doctors to choose specific inhibitors to treat diseases, we would strongly recommend that when possible, protein arrays be performed together with expression array and/or mutation analysis for patients with hematological malignancy. Additionally, microRNA, lncRNAs and epigenetic profiling of leukemic cells will also provide different perspectives for the specific characteristics of a particular evolving disease. Such technologies have been developed and now are widely accessible and affordable with less patient material. We believe that the accumulation of such data will help researchers to better design targeted drugs and clinicians to better treat patients with unique malignancy signature.
Acknowledgements
The authors want to thank the lab members for their contribution in preparation of the manuscript.
Funding: Funds from the Ministry of Science and Technology of China (2013BAI01B09, 2012CB966604) and from National Nature Science Foundation (81130074, 81470280) supported part of the studies and for publishing this manuscript.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell 2011;144:646-74. [Crossref] [PubMed]
- Shayesteh L, Lu Y, Kuo WL, et al. PIK3CA is implicated as an oncogene in ovarian cancer. Nat Genet 1999;21:99-102. [Crossref] [PubMed]
- Samuels Y, Wang Z, Bardelli A, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science 2004;304:554. [Crossref] [PubMed]
- Bachman KE, Argani P, Samuels Y, et al. The PIK3CA gene is mutated with high frequency in human breast cancers. Cancer Biol Ther 2004;3:772-5. [Crossref] [PubMed]
- Ikenoue T, Kanai F, Hikiba Y, et al. Functional analysis of PIK3CA gene mutations in human colorectal cancer. Cancer Res 2005;65:4562-7. [Crossref] [PubMed]
- Samuels Y, Velculescu VE. Oncogenic mutations of PIK3CA in human cancers. Cell Cycle 2004;3:1221-4. [Crossref] [PubMed]
- Bader AG, Kang S, Vogt PK. Cancer-specific mutations in PIK3CA are oncogenic in vivo. Proc Natl Acad Sci U S A 2006;103:1475-9. [Crossref] [PubMed]
- Fang WL, Huang KH, Lan YT, et al. Mutations in PI3K/AKT pathway genes and amplifications of PIK3CA are associated with patterns of recurrence in gastric cancers. Oncotarget 2016;7:6201-20. [PubMed]
- Shoji K, Oda K, Nakagawa S, et al. The oncogenic mutation in the pleckstrin homology domain of AKT1 in endometrial carcinomas. Br J Cancer 2009;101:145-8. [Crossref] [PubMed]
- Steck PA, Pershouse MA, Jasser SA, et al. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet 1997;15:356-62. [Crossref] [PubMed]
- Li J, Yen C, Liaw D, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science 1997;275:1943-7. [Crossref] [PubMed]
- Tashiro H, Blazes MS, Wu R, et al. Mutations in PTEN are frequent in endometrial carcinoma but rare in other common gynecological malignancies. Cancer Res 1997;57:3935-40. [PubMed]
- Wang SI, Puc J, Li J, et al. Somatic mutations of PTEN in glioblastoma multiforme. Cancer Res 1997;57:4183-6. [PubMed]
- Samuels Y, Ericson K. Oncogenic PI3K and its role in cancer. Curr Opin Oncol 2006;18:77-82. [Crossref] [PubMed]
- Dbouk HA, Khalil BD, Wu H, et al. Characterization of a tumor-associated activating mutation of the p110beta PI 3-kinase. PLoS One 2013;8:e63833. [Crossref] [PubMed]
- Cheung LW, Hennessy BT, Li J, et al. High frequency of PIK3R1 and PIK3R2 mutations in endometrial cancer elucidates a novel mechanism for regulation of PTEN protein stability. Cancer Discov 2011;1:170-85. [Crossref] [PubMed]
- Jaiswal BS, Janakiraman V, Kljavin NM, et al. Somatic mutations in p85alpha promote tumorigenesis through class IA PI3K activation. Cancer Cell 2009;16:463-74. [Crossref] [PubMed]
- Vanhaesebroeck B, Guillermet-Guibert J, Graupera M, et al. The emerging mechanisms of isoform-specific PI3K signalling. Nat Rev Mol Cell Biol 2010;11:329-41. [Crossref] [PubMed]
- Jaber N, Dou Z, Chen JS, et al. Class III PI3K Vps34 plays an essential role in autophagy and in heart and liver function. Proc Natl Acad Sci U S A 2012;109:2003-8. [Crossref] [PubMed]
- Chu EC, Tarnawski AS. PTEN regulatory functions in tumor suppression and cell biology. Med Sci Monit 2004;10:RA235-41. [PubMed]
- Alessi DR, James SR, Downes CP, et al. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Balpha. Curr Biol 1997;7:261-9. [Crossref] [PubMed]
- Manning BD, Cantley LC. AKT/PKB signaling: navigating downstream. Cell 2007;129:1261-74. [Crossref] [PubMed]
- Maurer M, Su T, Saal LH, et al. 3-Phosphoinositide-dependent kinase 1 potentiates upstream lesions on the phosphatidylinositol 3-kinase pathway in breast carcinoma. Cancer Res 2009;69:6299-306. [Crossref] [PubMed]
- Choucair KA, Guerard KP, Ejdelman J, et al. The 16p13.3 (PDPK1) genomic gain in prostate cancer: a potential role in disease progression. Transl Oncol 2012;5:453-60. [Crossref] [PubMed]
- Yang Z, Wu Z, Liu T, et al. Upregulation of PDK1 associates with poor prognosis in esophageal squamous cell carcinoma with facilitating tumorigenicity in vitro. Med Oncol 2014;31:337. [Crossref] [PubMed]
- Scortegagna M, Ruller C, Feng Y, et al. Genetic inactivation or pharmacological inhibition of Pdk1 delays development and inhibits metastasis of Braf(V600E)::Pten(-/-) melanoma. Oncogene 2014;33:4330-9. [Crossref] [PubMed]
- Zabkiewicz J, Pearn L, Hills RK, et al. The PDK1 master kinase is over-expressed in acute myeloid leukemia and promotes PKC-mediated survival of leukemic blasts. Haematologica 2014;99:858-64. [Crossref] [PubMed]
- Gagliardi PA, di Blasio L, Primo L. PDK1: A signaling hub for cell migration and tumor invasion. Biochim Biophys Acta 2015;1856:178-88.
- Staal SP. Molecular cloning of the akt oncogene and its human homologues AKT1 and AKT2: amplification of AKT1 in a primary human gastric adenocarcinoma. Proc Natl Acad Sci U S A 1987;84:5034-7. [Crossref] [PubMed]
- Cheng JQ, Ruggeri B, Klein WM, et al. Amplification of AKT2 in human pancreatic cells and inhibition of AKT2 expression and tumorigenicity by antisense RNA. Proc Natl Acad Sci U S A 1996;93:3636-41. [Crossref] [PubMed]
- Ruggeri BA, Huang L, Wood M, et al. Amplification and overexpression of the AKT2 oncogene in a subset of human pancreatic ductal adenocarcinomas. Mol Carcinog 1998;21:81-6. [Crossref] [PubMed]
- Bellacosa A, de Feo D, Godwin AK, et al. Molecular alterations of the AKT2 oncogene in ovarian and breast carcinomas. Int J Cancer 1995;64:280-5. [Crossref] [PubMed]
- Stahl JM, Sharma A, Cheung M, et al. Deregulated Akt3 activity promotes development of malignant melanoma. Cancer Res 2004;64:7002-10. [Crossref] [PubMed]
- Cristiano BE, Chan JC, Hannan KM, et al. A specific role for AKT3 in the genesis of ovarian cancer through modulation of G(2)-M phase transition. Cancer Res 2006;66:11718-25. [Crossref] [PubMed]
- Lin HP, Lin CY, Huo C, et al. AKT3 promotes prostate cancer proliferation cells through regulation of Akt, B-Raf, and TSC1/TSC2. Oncotarget 2015;6:27097-112. [Crossref] [PubMed]
- Carpten JD, Faber AL, Horn C, et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature 2007;448:439-44. [Crossref] [PubMed]
- Hashimoto K, Mori N, Tamesa T, et al. Analysis of DNA copy number aberrations in hepatitis C virus-associated hepatocellular carcinomas by conventional CGH and array CGH. Mod Pathol 2004;17:617-22. [Crossref] [PubMed]
- Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 2008;455:1061-8. [Crossref] [PubMed]
- Kaestner KH, Knochel W, Martinez DE. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev 2000;14:142-6. [PubMed]
- Fu Z, Tindall DJ. FOXOs, cancer and regulation of apoptosis. Oncogene 2008;27:2312-9. [Crossref] [PubMed]
- Greer EL, Brunet A. FOXO transcription factors at the interface between longevity and tumor suppression. Oncogene 2005;24:7410-25. [Crossref] [PubMed]
- Inoki K, Li Y, Xu T, et al. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev 2003;17:1829-34. [Crossref] [PubMed]
- Zhang Y, Gao X, Saucedo LJ, et al. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat Cell Biol 2003;5:578-81. [Crossref] [PubMed]
- Neuman NA, Henske EP. Non-canonical functions of the tuberous sclerosis complex-Rheb signalling axis. EMBO Molecular Medicine 2011;3:189-200. [Crossref] [PubMed]
- Kenerson HL, Aicher LD, True LD, et al. Activated mammalian target of rapamycin pathway in the pathogenesis of tuberous sclerosis complex renal tumors. Cancer Res 2002;62:5645-50. [PubMed]
- Zheng H, Liu A, Liu B, et al. Ras homologue enriched in brain is a critical target of farnesyltransferase inhibitors in non-small cell lung cancer cells. Cancer Lett 2010;297:117-25. [Crossref] [PubMed]
- Sun B, Karin M. Obesity, inflammation, and liver cancer. J Hepatol 2012;56:704-13. [Crossref] [PubMed]
- Platt FM, Hurst CD, Taylor CF, et al. Spectrum of phosphatidylinositol 3-kinase pathway gene alterations in bladder cancer. Clin Cancer Res 2009;15:6008-17. [Crossref] [PubMed]
- Kobayashi T, Shimizu Y, Terada N, et al. Regulation of androgen receptor transactivity and mTOR-S6 kinase pathway by Rheb in prostate cancer cell proliferation. Prostate 2010;70:866-74. [PubMed]
- Eom M, Han A, Yi SY, et al. RHEB expression in fibroadenomas of the breast. Pathol Int 2008;58:226-32. [Crossref] [PubMed]
- Gao Y, Gao J, Li M, et al. Rheb1 promotes tumor progression through mTORC1 in MLL-AF9-initiated murine acute myeloid leukemia. J Hematol Oncol 2016;9:36. [Crossref] [PubMed]
- Lu ZH, Shvartsman MB, Lee AY, et al. Mammalian target of rapamycin activator RHEB is frequently overexpressed in human carcinomas and is critical and sufficient for skin epithelial carcinogenesis. Cancer Res 2010;70:3287-98. [Crossref] [PubMed]
- Lawrence MS, Stojanov P, Mermel CH, et al. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature 2014;505:495-501. [Crossref] [PubMed]
- Laplante M, Sabatini DM. mTOR signaling. Cold Spring Harb Perspect Biol 2012.4. [PubMed]
- Manning G, Whyte DB, Martinez R, et al. The protein kinase complement of the human genome. Science 2002;298:1912-34. [Crossref] [PubMed]
- Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell 2012;149:274-93. [Crossref] [PubMed]
- Wang X, Chu Y, Wang W, et al. mTORC signaling in hematopoiesis. Int J Hematol 2016;103:510-8. [Crossref] [PubMed]
- Hoshii T, Matsuda S, Hirao A. Pleiotropic roles of mTOR complexes in haemato-lymphopoiesis and leukemogenesis. J Biochem 2014;156:73-83. [Crossref] [PubMed]
- Zhou F, Li X, Wang W, et al. Tracing haematopoietic stem cell formation at single-cell resolution. Nature 2016;533:487-92. [Crossref] [PubMed]
- Zick Y. Ser/Thr phosphorylation of IRS proteins: a molecular basis for insulin resistance. Sci STKE 2005;2005:pe4.
- Min YH, Eom JI, Cheong JW, et al. Constitutive phosphorylation of Akt/PKB protein in acute myeloid leukemia: its significance as a prognostic variable. Leukemia 2003;17:995-7. [Crossref] [PubMed]
- Kornblau SM, Tibes R, Qiu YH, et al. Functional proteomic profiling of AML predicts response and survival. Blood 2009;113:154-64. [Crossref] [PubMed]
- Chen W, Drakos E, Grammatikakis I, et al. mTOR signaling is activated by FLT3 kinase and promotes survival of FLT3-mutated acute myeloid leukemia cells. Mol Cancer 2010;9:292. [Crossref] [PubMed]
- Silva A, Yunes JA, Cardoso BA, et al. PTEN posttranslational inactivation and hyperactivation of the PI3K/Akt pathway sustain primary T cell leukemia viability. J Clin Invest 2008;118:3762-74. [Crossref] [PubMed]
- Kharas MG, Deane JA, Wong S, et al. Phosphoinositide 3-kinase signaling is essential for ABL oncogene-mediated transformation of B-lineage cells. Blood 2004;103:4268-75. [Crossref] [PubMed]
- Niemann CU, Jones J, Wiestner A. Towards targeted therapy of chronic lymphocytic leukemia. Adv Exp Med Biol 2013;792:259-91. [Crossref] [PubMed]
- Hyun T, Yam A, Pece S, et al. Loss of PTEN expression leading to high Akt activation in human multiple myelomas. Blood 2000;96:3560-8. [PubMed]
- Hu L, Shi Y, Hsu JH, et al. Downstream effectors of oncogenic ras in multiple myeloma cells. Blood 2003;101:3126-35. [Crossref] [PubMed]
- Nyåkern M, Tazzari PL, Finelli C, et al. Frequent elevation of Akt kinase phosphorylation in blood marrow and peripheral blood mononuclear cells from high-risk myelodysplastic syndrome patients. Leukemia 2006;20:230-8. [Crossref] [PubMed]
- Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer 2009;9:550-62. [Crossref] [PubMed]
- Fruman DA, Rommel C. PI3K and cancer: lessons, challenges and opportunities. Nat Rev Drug Discov 2014;13:140-56. [Crossref] [PubMed]
- Rodrik-Outmezguine VS, Okaniwa M, Yao Z, et al. Overcoming mTOR resistance mutations with a new-generation mTOR inhibitor. Nature 2016;534:272-6. [PubMed]
- Fransecky L, Mochmann LH, Baldus CD. Outlook on PI3K/AKT/mTOR inhibition in acute leukemia. Mol Cell Ther 2015;3:2. [Crossref] [PubMed]
- Casanova I, Bosch R, Lasa A, et al. A celecoxib derivative inhibits focal adhesion signaling and induces caspase-8-dependent apoptosis in human acute myeloid leukemia cells. Int J Cancer 2008;123:217-26. [Crossref] [PubMed]
- Feldman RI, Wu JM, Polokoff MA, et al. Novel small molecule inhibitors of 3-phosphoinositide-dependent kinase-1. J Biol Chem 2005;280:19867-74. [Crossref] [PubMed]
- Hu T, Li C, Zhang Y, et al. Phosphoinositide-dependent kinase 1 regulates leukemia stem cell maintenance in MLL-AF9-induced murine acute myeloid leukemia. Biochem Biophys Res Commun 2015;459:692-8. [Crossref] [PubMed]
- Bressanin D, Evangelisti C, Ricci F, et al. Harnessing the PI3K/Akt/mTOR pathway in T-cell acute lymphoblastic leukemia: eliminating activity by targeting at different levels. Oncotarget 2012;3:811-23. [Crossref] [PubMed]
- Tamburini J, Chapuis N, Bardet V, et al. Mammalian target of rapamycin (mTOR) inhibition activates phosphatidylinositol 3-kinase/Akt by up-regulating insulin-like growth factor-1 receptor signaling in acute myeloid leukemia: rationale for therapeutic inhibition of both pathways. Blood 2008;111:379-82. [Crossref] [PubMed]
- Tamburini J, Green AS, Bardet V, et al. Protein synthesis is resistant to rapamycin and constitutes a promising therapeutic target in acute myeloid leukemia. Blood 2009;114:1618-27. [Crossref] [PubMed]
- Medvetz D, Priolo C, Henske EP. Therapeutic targeting of cellular metabolism in cells with hyperactive mTORC1: a paradigm shift. Mol Cancer Res 2015;13:3-8. [Crossref] [PubMed]
- Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6:pl1. [Crossref] [PubMed]
- Ismail SI, Mahmoud IS, Msallam MM, et al. Hotspot mutations of PIK3CA and AKT1 genes are absent in multiple myeloma. Leuk Res 2010;34:824-6. [Crossref] [PubMed]
- Chang H, Qi XY, Claudio J, et al. Analysis of PTEN deletions and mutations in multiple myeloma. Leuk Res 2006;30:262-5. [Crossref] [PubMed]
Cite this article as: Gao Y, Yuan CY, Yuan W. Will targeting PI3K/Akt/mTOR signaling work in hematopoietic malignancies? Stem Cell Investig 2016;3:31.


