Parentally imprinted genes regulate hematopoiesis—new evidence from the Dlk1–Gtl2 locus
Parental imprinting and embryogenesis
Genomic imprinting is an epigenetic process that leads to the expression of a small set of genes in a parental origin-specific manner so that they are expressed either from the maternal or the paternal set of chromosomes (1). So far, only ~100 genes have been identified as parentally imprinted, and it is well known that they play crucial roles in embryogenesis, fetal growth, the totipotent state of the zygote, and the pluripotency of developmentally early stem cells (1-3). Genomic imprinting is also the most important mechanism preventing parthenogenesis (4,5). Deregulation of the expression of imprinted genes during embryogenesis also leads to developmental disorders such as Beckwith-Wiedemann syndrome, Angelman syndrome, and Prader-Willi syndrome (1).
Most imprinted genes are organized into clusters that contain a region with differential methylation (methylated or unmethylated), which has different methylation patterns depending on the parental origin of the chromosome, i.e., from the mother or father. This DNA region, called a differentially methylated region (DMR), is the main epigenetic mark that controls appropriate monoallelic expression of imprinted genes (6). Interestingly, most DMRs are methylated on the maternal chromosome and only a few on the paternal chromosome, and it seems that these latter DMRs are highly important (7).
Among imprinted genes, two paternally imprinted loci that regulate expression of the tandem genes Igf2–H19 and Dlk1–Gtl2 are of special interest (Figure 1A). These tandem genes contain DMRs located between Igf2 and H19 and between Dlk1 and Gtl2, respectively. While Igf2 encodes insulin-like growth factor 2, Dlk1 encodes delta-like homolog 1, a cell-surface transmembrane protein belonging to the epidermal growth factor-like homeotic family. The other segments of the two tandem genes are the H19 and Gtl2 loci, which encode noncoding RNAs (ncRNA) that are processed into several miRNAs regulating the expression of many genes involved in cell proliferation and metabolism (8,9).
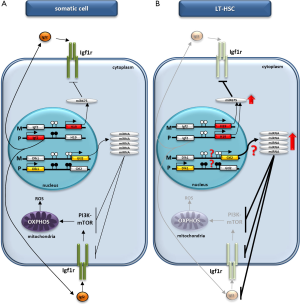
During embryogenesis, the DMRs in both the Igf2–H19 and Dlk1–Gtl2 loci become erased in migrating primordial germ cells (PGCs), which are precursor cells of gametes (10). This erasure results in downregulation of Igf2 and Dlk1 expression and upregulation of the ncRNAs transcribed from the H19 and Gtl2 loci in these cells, which are endowed with developmental totipotency. This gene regulation results in PGC quiescence and prevents them from teratoma formation. Proper reestablishment of a somatic type of imprint within the Igf2–H19 and Dlk1–Gtl2 loci in male gametes occurs after remethylation of the erased DMR after the first meiotic division, when the number of chromosomes in the precursors of sperm cells is reduced to the haploid number (11). Thus, at the time of fertilization, the DMRs within the Igf2–H19 and Dlk1–Gtl2 loci are remethylated on the paternal chromosomes; however, they stay unmethylated on the maternal chromosomes, which is the basis for the monoallelic expression from these loci, resulting in the balanced expression of growth-promoting Igf2 and Dlk1 genes and growth-suppressing H19 and Gtl2 genes (Figure 1A). The proper intracellular ratio of the products of both tandem genes is required for normal embryonic development.
Seminal studies describing the creation of viable bimaternal mice, which contain two maternal haploid sets of chromosomes from two oocytes (one non-growing and the other fully grown) demonstrated that proper expression of these two tandem genes is crucial for embryogenesis in the same way as in somatic cells (5,12). This was achieved by creating two oocytes (fertilized zygotes), with the DMR sequences for both the Igf2–H19 and Dlk1–Gtl2 loci deleted in one of the oocytes. This experimental procedure allows both tandem gene products to be expressed at a physiological ratio and mimics the situation encountered in normal zygotes, which are created after fertilization of the oocyte by the sperm. Moreover, by employing a bimaternal mouse model, it was demonstrated that both the Igf2 and Dlk1 genes are required for proper fetal liver (FL) hematopoiesis (13).
Parental imprinting and its newly recognized impact on adult stem cells
For many years the regulation of gene expression by epigenetic changes within DMRs was attributed to stem cells only during embryonic development. However, as we demonstrated almost a decade ago, these epigenetic changes also play a role in regulating the fate of postnatal stem cells, such as in a population of rare and quiescent pluripotent/multipotent stem cells known as very small embryonic-like stem cells (VSELs) isolated from murine bone marrow (BM) and FL (14,15). These cells are believed to be at the top of the stem cell hierarchy in adult tissues and in several set of experiments have been reported to give rise to hematopoietic stem cells (HSCs) (16). Based on their potential for hematopoietic specification, we proposed that VSELs are precursors of long-term repopulating HSCs (LT-HSCs).
Igf2-H19 locus as master regulator of HSCs fate
It has been demonstrated that VSELs freshly isolated from murine BM erase the paternally methylated imprints at regulatory DMRs, such as within the Igf2–H19 and Rasgrf1 loci. However, they also hypermethylate the maternally methylated imprints at DMRs for the Igf2 receptor (Igf2r), Kcnq1-p57KIP2, and Peg1 loci. Because paternally expressed imprinted genes (Igf2 and Rasgrf1) enhance embryonic growth, and maternally expressed genes (H19, p57KIP2, and Igf2r) inhibit cell proliferation, the unique genomic imprinting pattern observed in VSELs demonstrates the growth-repressive influence of imprinted genes, including the Igf2–H19 locus, on the proliferation of these cells. One miRNA derived from the H19 ncRNA has been demonstrated to inhibit expression of the signaling receptor for Igf2, the insulin-like growth factor 1 receptor (Igf1r) (8), and in addition we have proposed the same phenomenon for the insulin receptor itself (17). Therefore, VSELs are in a quiescent state because of attenuation of insulin/insulin-like growth factor signaling (18,19).
In line with this observation, Li and colleagues reported on a model of the maternal type of imprinting (mimicking erasure within the Igf2–H19 locus) that maintained the quiescence of murine LT-HSCs (20). In well-designed experiments involving several murine knockouts, it was demonstrated that conditional deletion of the maternal but not the paternal DMR for the Igf2–H19 locus reduces adult HSC quiescence, a state required for long-term maintenance of HSCs, and compromises HSC function (20). Thus, this work confirmed our results obtained with VSELs that epigenetic regulation of Igf2–H19 at the paternal chromosome, which is so critical for embryonic growth and development, operates also in regulating the quiescence of stem cells isolated from adult tissues. Again, as was proposed for VSELs (18-21), the Li group found that upregulation of H19 gives rise to miRNA-675, which suppresses Igf1r expression and inhibits Igf2–Igf1r signaling (20).
Dlk1-Meg3 locus as novel master regulator of HSCs fate
In a more recently published paper, the Li group also reported exciting results for another imprinted tandem gene, Dlk1–Gtl2, and its involvement in hematopoiesis and LT-HSC function (22). Apart from Dlk1, the Dlk1–Gtl2 locus also encodes two other proteins, Rtl1 and Dio3, which are both transcribed from the paternally inherited chromosome, whereas the maternally inherited chromosome is a source of the largest ncRNA mega-cluster in mammals, which gives rise to many different miRNAs (9). Besides the Ig2–H19 locus the Dlk1–Gtl2 locus was shown in the bimaternal mouse model to play a crucial role in embryonic and adult stem cell biology and FL hematopoiesis (12,13).
To better address the role of the Dlk1–Gtl2 locus in murine hematopoiesis, Qian et al. performed transcriptome profiling in 17 subpopulations of HSCs, progenitors, and mature lineages and found an ncRNA expression fingerprint unique to each subpopulation (22). Interestingly, the main enrichment in LT-HSCs was observed for ncRNAs transcribed from the Dlk1–Gtl2 locus. Based on this finding, the authors decided to better characterize the role of this region in maintaining the quiescent phenotype of LT-HSCs. By creating several knockout animals, they were able to show that deletion of the DMR for the Dlk1–Gtl2 locus or the Gtl2 gene on the maternal chromosome results in downregulation of a Gtl2-derived ncRNA and downstream miRNAs cluster, which results in activation of the PI3K–mTOR pathway and an increase in mitochondrial biogenesis and metabolic activity. The resulting enhanced ROS eventually leads to the apoptosis of HSCs. Moreover, the authors were able to confirm that this observed phenotype is not the result of overexpression of Dlk1 but the effect of downregulation of several miRNAs processed from the ncRNA transcribed from the Gtl2 locus.
Since some of the knockouts were lethal after day E16, several experiments were performed employing FL HSCs, which led to a further intriguing observation. Even though adult LT-HSCs, in contrast to fetal HSCs, were kept in a quiescent state, both FL HSCs and LT-HSCs shared a similar ncRNA expression pattern, especially with respect to ncRNAs transcribed from the Gtl2 locus. This finding suggests that Gtl2-derived ncRNAs play a role in maintaining both fetal and adult HSCs, and a manipulation that led to a decrease in Gtl2 expression resulted in an impaired long-term reconstitution capacity of HSCs by affecting their function through mitochondrial metabolic activation. To confirm the effect of miRNAs on the reconstitution capacity of HSCs, 10 highly expressed miRNAs from the Dlk1–Gtl2 locus were overexpressed in fetal HSCs from mice with a maternal deletion of the Dlk1–Gtl2 DMR and low expression of the Gtl2-derived miRNA. It has been observed that, in mice 16 weeks after transplantation of HSCs transduced with Gtl2-derived miRNAs, the absolute number of HSCs and their repopulation potential was higher than in mice transplanted with control HSCs. This suggests that miRNAs derived from the ncRNA transcribed from Gtl2 locus plays a role in the maintenance of LT-HSCs.
It is important to emphasize that there are similarities in the roles of ncRNAs transcribed from the Igf2–H19 and Dlk1–Gtl2 loci, as H19 ncRNA and Gtl2 ncRNA with downstream miRNA cluster give rise to miRNAs that target the Igf2–Igf1r signaling pathway, which emphasizes the predominant role of this axis in regulating the quiescent state of LT-HSCs (Figure 1B). The attenuation of this signaling has been proposed by us to be involved in maintaining the quiescent state of VSELs (18,19).
However, even though much work has been done by the Li group to characterize the role of the Gtl2–Dlk1 locus in maintaining LT-HSC function, one question still remains: what is the mechanism that regulates the increased expression of Gtl2-derived miRNAs in LT-HSCs? While the authors were able to show convincingly that imprinting within the Dlk1–Gtl2 locus is crucial in regulating hematopoiesis, unfortunately, they did not study in detail the differences in methylation state of the DMR at the Dlk1–Gtl2 locus between different subpopulations of hematopoietic cells that they analyzed, and perhaps these differences could explain the ncRNA fingerprints that differed between the cell populations employed in this interesting study.
Conclusions
Based on these intriguing results, we can expect more results in the near future addressing the role of imprinted genes in the regulatory biology of LT-HSCs as well as other types of adult stem cells. As an example, RasGrf1 is another interesting paternally imprinted gene candidate in mice which encodes GTP exchange factor (GEF) (23). The DMR for this gene has been found to be erased in VSELs isolated from postnatal BM (18). It would be interesting to address in more detail its role in regulating the biology of LT-HSCs, particularly because RasGrf1, like GEF protein, is involved in Igf1r and insulin receptor signaling.
Acknowledgements
Funding: This work was supported by NIH grants 2R01 DK074720 and R01HL112788, the Stella and Henry Endowment, and Harmonia NCN grant: UMO-2014/14/M/NZ3/00475 to MZ Ratajczak.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Peters J. The role of genomic imprinting in biology and disease: an expanding view. Nat Rev Genet 2014;15:517-30. [Crossref] [PubMed]
- Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat Rev Genet 2001;2:21-32. [Crossref] [PubMed]
- Yamazaki Y, Mann MR, Lee SS, et al. Reprogramming of primordial germ cells begins before migration into the genital ridge, making these cells inadequate donors for reproductive cloning. Proc Natl Acad Sci U S A 2003;100:12207-12. [Crossref] [PubMed]
- Kono T. Genomic imprinting is a barrier to parthenogenesis in mammals. Cytogenet Genome Res 2006;113:31-5. [Crossref] [PubMed]
- Kono T, Obata Y, Wu Q, et al. Birth of parthenogenetic mice that can develop to adulthood. Nature 2004;428:860-4. [Crossref] [PubMed]
- Ferguson-Smith AC. Genomic imprinting: the emergence of an epigenetic paradigm. Nat Rev Genet 2011;12:565-75. [Crossref] [PubMed]
- Geuns E, De Temmerman N, Hilven P, et al. Methylation analysis of the intergenic differentially methylated region of DLK1-GTL2 in human. Eur J Hum Genet 2007;15:352-61. [Crossref] [PubMed]
- Keniry A, Oxley D, Monnier P, et al. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol 2012;14:659-65. [Crossref] [PubMed]
- Seitz H, Royo H, Bortolin ML, et al. A large imprinted microRNA gene cluster at the mouse Dlk1-Gtl2 domain. Genome Res 2004;14:1741-8. [Crossref] [PubMed]
- Kelsey G, Feil R. New insights into establishment and maintenance of DNA methylation imprints in mammals. Philos Trans R Soc Lond B Biol Sci 2013;368:20110336. [Crossref] [PubMed]
- Sasaki H, Matsui Y. Epigenetic events in mammalian germ-cell development: reprogramming and beyond. Nat Rev Genet 2008;9:129-40. [Crossref] [PubMed]
- Kawahara M, Wu Q, Takahashi N, et al. High-frequency generation of viable mice from engineered bi-maternal embryos. Nat Biotechnol 2007;25:1045-50. [Crossref] [PubMed]
- Wu Q, Kawahara M, Kono T. Synergistic role of Igf2 and Dlk1 in fetal liver development and hematopoiesis in bi-maternal mice. J Reprod Dev 2008;54:177-82. [Crossref] [PubMed]
- Zuba-Surma EK, Kucia M, Rui L, et al. Fetal liver very small embryonic/epiblast like stem cells follow developmental migratory pathway of hematopoietic stem cells. Ann N Y Acad Sci 2009;1176:205-18. [Crossref] [PubMed]
- Kucia M, Reca R, Campbell FR, et al. A population of very small embryonic-like (VSEL) CXCR4(+)SSEA-1(+)Oct-4+ stem cells identified in adult bone marrow. Leukemia 2006;20:857-69. [Crossref] [PubMed]
- Ratajczak J, Wysoczynski M, Zuba-Surma E, et al. Adult murine bone marrow-derived very small embryonic-like stem cells differentiate into the hematopoietic lineage after coculture over OP9 stromal cells. Exp Hematol 2011;39:225-37. [Crossref] [PubMed]
- Tarnowski M, Tkacz M, Czerewaty M, et al. 5-Azacytidine inhibits human rhabdomyosarcoma cell growth by downregulating insulin-like growth factor 2 expression and reactivating the H19 gene product miR-675, which negatively affects insulin-like growth factors and insulin signaling. Int J Oncol 2015;46:2241-50. [PubMed]
- Shin DM, Zuba-Surma EK, Wu W, et al. Novel epigenetic mechanisms that control pluripotency and quiescence of adult bone marrow-derived Oct4(+) very small embryonic-like stem cells. Leukemia 2009;23:2042-51. [Crossref] [PubMed]
- Maj M, Schneider G, Ratajczak J, et al. The cell cycle- and insulin-signaling-inhibiting miRNA expression pattern of very small embryonic-like stem cells contributes to their quiescent state. Exp Biol Med (Maywood) 2015;240:1107-11. [Crossref] [PubMed]
- Venkatraman A, He XC, Thorvaldsen JL, et al. Maternal imprinting at the H19-Igf2 locus maintains adult haematopoietic stem cell quiescence. Nature 2013;500:345-9. [Crossref] [PubMed]
- Ratajczak MZ, Shin DM, Schneider G, et al. Parental imprinting regulates insulin-like growth factor signaling: a Rosetta Stone for understanding the biology of pluripotent stem cells, aging and cancerogenesis. Leukemia 2013;27:773-9. [Crossref] [PubMed]
- Qian P, He XC, Paulson A, et al. The Dlk1-Gtl2 locus preserves LT-HSC function by inhibiting the PI3K-mTOR pathway to restrict mitochondrial metabolism. Cell Stem Cell 2016;18:214-28. [Crossref] [PubMed]
- Fernández-Medarde A, Santos E. The RasGrf family of mammalian guanine nucleotide exchange factors. Biochim Biophys Acta 2011;1815:170-88.
Cite this article as: Schneider G, Sellers ZP, Ratajczak MZ. Parentally imprinted genes regulate hematopoiesis—new evidence from the Dlk1–Gtl2 locus. Stem Cell Investig 2016;3:29.


