Direct reprogramming to multipotent trophoblast stem cells, and is pluripotency needed for regenerative medicine either?
For the clinical applications of regenerative medicine, induced pluripotent stem cells (iPSCs) (all acronyms are defined in the Glossary at the end of the text) have advantages over embryonic stem cells (ESCs) in histocompatibility and the lack of reliance on embryo derivation. Induction of pluripotency can be achieved by transiently expressing a minimal set of transcription factors (TFs) to reprogram differentiated cells. Direct reprogramming of somatic cells into progenitor stem or other mature cells would eliminate the need to make iPSCs, and then direct differentiation and select the desired lineage. Another advantage of direct reprogramming is that by avoiding the pluripotent step, one mitigates the risk of tumorigenicity. Direct reprogramming requires knowledge of the reprogramming factors and knowledge of the culture conditions necessary to maintain the induced multipotent stem cells. The culture conditions for mouse trophoblast stem cells (TSCs) were defined in the 1990s (1,2). Knowledge of how to reprogram mouse fibroblast cells to multipotent TSC has now been provided by two recent publications from the Buganim (3) and the Schorle labs (4). These studies inform not only regenerative medicine but will also aid our understanding of normal and pathological placental development in early pregnancy.
More than 12 TFs related to TSC maintenance or reprogramming in general were transiently expressed and tested for their ability to induce TSC from mouse fibroblasts (3,4). Table 1 lists the induction and proof of validity of the induced TSC (iTSC) in each report; comparing the reprogramming, maintenance, and differentiation of iTSC in vitro and in vivo, as well as the transcriptome and epigenetic modification of iTSC compared with blastocyst-derived TSC (bdTSC).
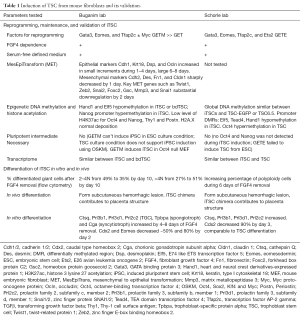
Full table
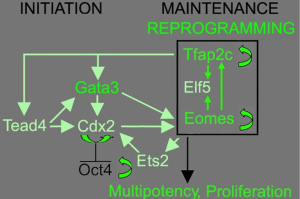
Of the TFs tested, three were identified in both reports as necessary for successful TSC induction, which were GATA Binding Protein 3 (Gata3), Eomesodermin (Eomes), and Transcription factor AP-2 gamma (Tfap2c). Surprisingly, genes like TEA domain transcription factor 4 (Tead4), E74 like ETS transcription factor 5 (Elf5) and caudal type homeobox 2 (Cdx2), which are known to be essential in TSC initiation and maintenance were not among them (Figure 1). Eomes and Tfap2c bind to Tead4, Gata3 and Elf5 genes, and positively regulate the expression of Elf5 (5), which is gatekeeper gene in trophoblast lineage differentiation in early blastocyst (6). Triple occupancy of Elf5, Eomes and Tfap2c were enriched in genes related to TSC proliferation and potency, including themselves (7). Gata3 binds to and positively regulates the expression Cdx2 (8), which increases the expression of itself as well as Elf5 and Eomes (5). Gata3 also induces the expression of Eomes independent of Cdx2 (9). E26 avian leukemia oncogene 2 (Ets2) is downstream of Eomes and directly regulates the transcription of Cdx2 (10). Eomes, Tfap2c, Gata3 and Ets2 function coordinately to activate their own endogenous expression and the expression of other genes involved in TSC initiation and maintenance, thus forming a self-reinforcing auto-regulatory circuitry. Ets2 was found to exist in 4 of the 5 genotyped iTSC clones by the Buganim lab (3), suggesting that Ets2 may promote successful reprogramming. The Schorle lab (4) identified Ets2 as one of the four TFs necessary for TSC induction. Note that the Schorle lab (4) used a shorter induction time of 10 days instead of the 20 days used by the Buganim lab (3), which may explain the difference in the necessity of Ets2. As with bdTSC, iTSC in both studies can be maintained in vitro with mouse embryonic fibroblast conditioned medium supplemented with fibroblast growth factor 4 (FGF4) or chemically defined TX medium supplemented with transforming growth factor β (TGF-β) (11).
TSC grow as epithelium and if epithelium formation is disturbed, potency is lost and differentiation occurs (12). Interestingly, although TFs that maintain mesenchymal state were lost in the first 2 days, it took 6–8 days to activate epithelial markers (Table 1). Follow-up studies should test for the kinetics of TSC-specific mRNA and epigenetic change during induction.
Reprogramming involves significant epigenetic changes, and it is important that the reprogrammed cell type resembles its naturally derived equivalent epigenetically. Fibroblast-derived iTSC have similar epigenetic profiles compared with bdTSC, as opposed to TSC differentiated in vitro from ESC, which is not complete largely due to epigenetic barriers (13). Both groups showed hypomethylation and activation of the TSC gatekeeper gene Elf5 and hypermethylation of ESC lineage markers octamer-binding transcription factor 4 (Oct4) or Nanog. Global DNA methylation signatures of iTSC are comparable to bdTSC (4). Histone acetylation analysis further supports the successful induction of iTSC. Histone 3 lysine 27 acetylation (H3K27ac) marks accessible chromatin for transcription. TSC-specific gene regions were more enriched with H3K27ac in iTSC, while genes related to ESC such as Nanog and Oct4, or fibroblast such as Thy1 and periostin (Postn, aka osteoblast Specific Factor) showed lower H3K27ac levels (Table 1). In addition, genome-wide deposition pattern of histone variant H2A.X in iTSC resembles that in bdTSC (3). Together, they showed that iTSC are comparable to iTSC epigenetically.
Further evidence of the validity of iTSC includes its differentiation potential both in vitro and in vivo. During in vitro differentiation of iTSC, the loss of potency factor Cdx2 and Eomes and gain of differentiation factors such as trophoblast specific protein alpha (Tpbpa), prolactin family 3, subfamily d, member 1 (Prl3d1) (Table 1) resembled bdTSC both in chronological sequence and magnitude. However, although >50% TSC differentiated to >4N giant cells, over a third of 2–4N cells remained 10 days after FGF4 removal. Whether the 2–4N cells were terminally differentiated or differentiating cells with limited proliferation potential remains unknown. Similar to the concern of tumorigenicity in iPSC-based therapy, it can be a potential problem if these cells continue to proliferate in differentiation milieu in vivo. Upon subcutaneous injection into nude mice, iTSC formed transient hemorrhagic lesions resembling invading trophoblast giant cells observed during implantation. iTSC also contributed to placental structures when injected into blastocyst. However, iTSC induced by the TF cocktail lacking Ets2 in Buganim lab’s report (3) seemed to have lower contribution to developing placenta.
Both groups showed that iTSC induction was not associated with iPSC intermediate stage. The less successful efforts to differentiate ESC into TSC further supports direct TSC reprogramming. The supposed proliferative advantage of iPSCs over direct reprogramming is questionable, when the efficiency of subsequent differentiation and selection is taken into consideration. And there is proliferative potential in some directly reprogrammed multipotent stem cells, variable though that may be. Direct reprogramming faces similar efficacy and safety issues as the two-step iPSC induction and directed differentiation. But the encouraging results of iTSC induction from fibroblasts indicate there is no monopoly on ideas of how to best proceed with reprogramming and regenerative medicine.
Acknowledgements
Funding: This research was supported by grants to DA Rappolee from NIH (1R03HD061431) and from the Office of the Vice President for Research at Wayne State University, and support for GC Parker from NIH (P30 ES020957).
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Comment on: Kubaczka C, Senner CE, Cierlitza M, et al. Direct induction of trophoblast stem cells from murine fibroblasts. Cell Stem Cell 2015;17:557-68.
Benchetrit H, Herman S, van Wietmarschen N, et al. Extensive nuclear reprogramming underlies lineage conversion into functional trophoblast stem-like cells. Cell Stem Cell 2015;17:543-56.
References
- Chai N, Patel Y, Jacobson K, et al. FGF is an essential regulator of the fifth cell division in preimplantation mouse embryos. Dev Biol 1998;198:105-15. [Crossref] [PubMed]
- Tanaka S, Kunath T, Hadjantonakis AK, et al. Promotion of trophoblast stem cell proliferation by FGF4. Science 1998;282:2072-5. [Crossref] [PubMed]
- Benchetrit H, Herman S, van Wietmarschen N, et al. Extensive nuclear reprogramming underlies lineage conversion into functional trophoblast stem-like cells. Cell Stem Cell 2015;17:543-56. [Crossref] [PubMed]
- Kubaczka C, Senner CE, Cierlitza M, et al. Direct induction of trophoblast stem cells from murine fibroblasts. Cell Stem Cell 2015;17:557-68. [Crossref] [PubMed]
- Kidder BL, Palmer S. Examination of transcriptional networks reveals an important role for TCFAP2C, SMARCA4, and EOMES in trophoblast stem cell maintenance. Genome Res 2010;20:458-72. [Crossref] [PubMed]
- Ng RK, Dean W, Dawson C, et al. Epigenetic restriction of embryonic cell lineage fate by methylation of Elf5. Nat Cell Biol 2008;10:1280-90. [Crossref] [PubMed]
- Latos PA, Sienerth AR, Murray A, et al. Elf5-centered transcription factor hub controls trophoblast stem cell self-renewal and differentiation through stoichiometry-sensitive shifts in target gene networks. Genes Dev 2015;29:2435-48. [Crossref] [PubMed]
- Home P, Ray S, Dutta D, et al. GATA3 is selectively expressed in the trophectoderm of peri-implantation embryo and directly regulates Cdx2 gene expression. J Biol Chem 2009;284:28729-37. [Crossref] [PubMed]
- Ralston A, Cox BJ, Nishioka N, et al. Gata3 regulates trophoblast development downstream of Tead4 and in parallel to Cdx2. Development 2010;137:395-403. [Crossref] [PubMed]
- Wen F, Tynan JA, Cecena G, et al. Ets2 is required for trophoblast stem cell self-renewal. Dev Biol 2007;312:284-99. [Crossref] [PubMed]
- Kubaczka C, Senner C, Araúzo-Bravo MJ, et al. Derivation and maintenance of murine trophoblast stem cells under defined conditions. Stem Cell Reports 2014;2:232-42. [Crossref] [PubMed]
- Abell AN, Granger DA, Johnson NL, et al. Trophoblast stem cell maintenance by fibroblast growth factor 4 requires MEKK4 activation of Jun N-terminal kinase. Mol Cell Biol 2009;29:2748-61. [Crossref] [PubMed]
- Cambuli F, Murray A, Dean W, et al. Epigenetic memory of the first cell fate decision prevents complete ES cell reprogramming into trophoblast. Nat Commun 2014;5:5538. [Crossref] [PubMed]
Cite this article as: Yang Y, Parker GC, Puscheck EE, Rappolee DA. Direct reprogramming to multipotent trophoblast stem cells, and is pluripotency needed for regenerative medicine either? Stem Cell Investig 2016;3:24.


