Cohesin complex is a major player on the stage of leukemogenesis
Epigenetic changes are increasingly being recognized as part of the multistep tumorigenesis process in cancer. These changes, including DNA methylation, covalent histone modifications, and nucleosome remodeling, work together to regulate the functioning of the genome by altering the local structural dynamics of chromatin, primarily regulating its accessibility and compactness (1). Other chromatin organizing factors are believed to bind to noncoding regions and control long-range looping of DNA, which is important in regulating the spatial relationships between gene enhancers and promoters that control gene expression (2).
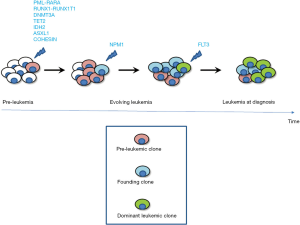
Epigenetic changes are pivotal for the understanding of the molecular events underlying leukemogenesis and more specifically the pathophysiology of acute leukemias, which despite being a diverse group of hematopoietic neoplasias, share a common pathogenetic theme. In fact, it is now widely accepted that mutations serially accumulate in clones of self-renewing hematopoietic stem and progenitor cells (HSPCs) before the disease arises abruptly and the clinical signs develop. Some mutations occur in pre-leukemic clones and in genes known to regulate epigenome, including cohesin (3,4). In particular the cohesin complex is involved in three processes: (I) maintenance of the polarity of sister chromatids during mitosis; (II) repair of double-strand DNA damage; and (III) regulation of transcription trough transcriptional factor (TF) recruitment and its interaction with CCCTC-binding factor (CTCF) (5). The complex functions to hold chromatin strands within a ring-like structure composed of the four core components RAD21, STAG2, SMC1A and SMC3 (6). Interestingly, mutations in the cohesin complex have been found in a variety of human neoplasms, and especially in myeloid malignancies (7). Figure 1 depicts schematically the multiphase process involved in acute myeloid leukemia (AML), with chromosomal translocations and key genes known to be mutated at each phase (for a recent comprehensive review on this subject see Grimwade et al. (8).
In order to understand the role of the cohesin complex in leukemogenesis, Mullenders and colleagues (9) have created RNA interference mouse models that inducibly knock down RAD21, SMC1A, and STAG2 systemically. Instead to ablate completely the expression of these genes, they decided to suppress cohesin transcripts and to reduce cohesin protein expression. The induction of the short hairpin RNA (shRNA)-mediated interference in vitro and in vivo resulted in a shift in the hematopoietic stem compartment towards myeloid lineages and increased the replating capacity of these cells. Over time, cohesin knockdown mice developed clinical features of myeloproliferative disorders/neoplasms. Moreover, a genome-wide mapping of chromatin accessibility on Lineage−Sca1+c-kit+ (LSK) cells obtained from shRNA mouse models allowed to establish that myeloid-associated genes were highly accessible in these mice. Further genomic analysis proved that the most enriched transcription factors motif is the GATA factor motif, GATA1 being a master regulator of primitive and definitive hematopoiesis.
Mazumdar and colleagues (10) have recently confirmed and extended these results, which demonstrate the role of cohesin protein complex mutants in hematopoiesis, especially in HSPC maintenance. They were able to show that missense and nonsense mutations in RAD21, STAG2, SMC1A, and SMC3 overexpression in AML cell lines and primary cord blood-derived human HSPCs resulted in a differentiation block with an increase frequency of CD34+ progenitor cells. Mutant cohesin increased the serial replating ability of human HSPCs in vitro and showed enrichment for HSCs and leukemia stem cell gene expression programs, indicating an effect to enforce stem cell functions. Mutant cohesin seems to impart its differentiation block only in the most immature HSCs and multipotent progenitors (MMPs), this finding being compatible with the often identified cohesin mutations in pre-leukemic HSCs in human AML (3,11). Interestingly, a skewed differentiation toward the myeloid lineage was observed both in vitro and in vivo.
In order to get detailed information on the underlying mechanism, the chromatin accessibility was investigated. Using ATAC-seq (assay for transposase-accessible chromatin followed by sequencing), they determined that mutant cohesin lead to a state of global loss of open chromatin and to an increase in accessibility at specific motifs for key hematopoietic TFs ERG, GATA2, and RUNX1 that in turn execute stem cell transcriptional programs and phenotypes. This notion was supported by the reverting effect on the mutant cohesin-induced differentiation block exerted by the knockdown of these three TFs. Overall, these data, together with those previously published (3,4), support the notion that pre-leukemic clones can arise due to occurring mutations in genes involved in global chromatin changes such as DNA methylation, histone modification, and chromatin looping (including cohesin).
Notably, the genetic complexity of leukemias represents a huge challenge to successful translation into clinical practice and many genes are recurrently mutated in AML. Furthermore, each leukemia case harbours multiple mutations and is potentially composed of subclones with differing mutational composition. Therefore it is important to identify critical AML-defining molecular abnormalities that distinguish real disease entities and mutations that have been demonstrated to confer prognostic information or that are therapeutically relevant.
In this light there are still many issues about the cohesin complex which should be addressed in the near future. How is the cohesin complex role integrated at the level of pre-leukemic clones with the other “landscaping” genes? Can the cohesin complex mutations be found during remission and prepare the stage for relapse? Is the cohesin complex a good target for drug design? In regard to the latter issue, epigenetic changes are more easily targetable than genetic changes, as enzymes and protein complexes are generally good drug targets. A few drugs targeting epigenetic changes in AML and other leukemias have already reached the clinical arena (12). Several other targeted therapies are needed in order to cure AML patients, and the identification of cohesin as a key protein complex in leukemogenesis could certainly contribute to the development of new drugs that specifically target mutant stem cells. From the work by Mazumdar et al. (10) it seems that cancer stem cells are those where these events primarily occur, and thus novel therapies should address this cellular compartment. Indeed, the cohesin complex is likely to be a good target in view of the fact that its down-regulation does not bring to any genomic instability (9), presumably not leading to unwanted dangerous side effects.
Hopefully, in the near future we will be able to use such molecular information to further improve diagnosis, identify subsets of patients eligible for novel targeted therapies, refine outcome prediction, and obtain significant treatment response.
Acknowledgements
This work was supported in part by Associazione Italiana contro Leucemie-Linfomi e Mieloma (AIL) Foggia.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis 2010;31:27-36. [Crossref] [PubMed]
- Hübner MR, Eckersley-Maslin MA, Spector DL. Chromatin organization and transcriptional regulation. Curr Opin Genet Dev 2013;23:89-95. [Crossref] [PubMed]
- Corces-Zimmerman MR, Hong WJ, Weissman IL, et al. Preleukemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proc Natl Acad Sci U S A 2014;111:2548-53. [Crossref] [PubMed]
- Corces-Zimmerman MR, Majeti R. Pre-leukemic evolution of hematopoietic stem cells: the importance of early mutations in leukemogenesis. Leukemia 2014;28:2276-82. [Crossref] [PubMed]
- Panigrahi AK, Pati D. Higher-order orchestration of hematopoiesis: is cohesin a new player? Exp Hematol 2012;40:967-73. [Crossref] [PubMed]
- Díaz-Martínez LA, Clarke DJ. Chromosome cohesion and the spindle checkpoint. Cell Cycle 2009;8:2733-40. [Crossref] [PubMed]
- Leiserson MD, Vandin F, Wu HT, et al. Pan-cancer network analysis identifies combinations of rare somatic mutations across pathways and protein complexes. Nat Genet 2015;47:106-14. [Crossref] [PubMed]
- Grimwade D, Ivey A, Huntly BJ. Molecular landscape of acute myeloid leukemia in younger adults and its clinical relevance. Blood 2016;127:29-41. [Crossref] [PubMed]
- Mullenders J, Aranda-Orgilles B, Lhoumaud P, et al. Cohesin loss alters adult hematopoietic stem cell homeostasis, leading to myeloproliferative neoplasms. J Exp Med 2015;212:1833-50. [Crossref] [PubMed]
- Mazumdar C, Shen Y, Xavy S, et al. Leukemia-associated cohesin mutants dominantly enforce stem cell programs and impair human hematopoietic progenitor differentiation. Cell Stem Cell 2015;17:675-88. [Crossref] [PubMed]
- Jan M, Snyder TM, Corces-Zimmerman MR, et al. Clonal evolution of preleukemic hematopoietic stem cells precedes human acute myeloid leukemia. Sci Transl Med 2012;4:149ra118. [Crossref] [PubMed]
- Graubert TA, Brunner AM, Fathi AT. New molecular abnormalities and clonal architecture in AML: from reciprocal translocations to whole-genome sequencing. Am Soc Clin Oncol Educ Book 2014.e334-40. [Crossref] [PubMed]
Cite this article as: Conese M, Liso A. Cohesin complex is a major player on the stage of leukemogenesis. Stem Cell Investig 2016;3:18.


